2OVR
 
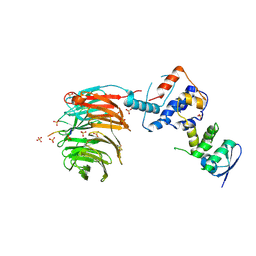 | | Structure of the Skp1-Fbw7-CyclinEdegN complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
2OVQ
 
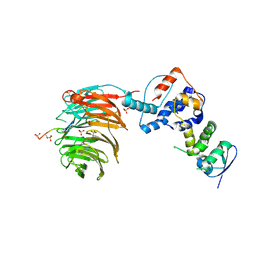 | | Structure of the Skp1-Fbw7-CyclinEdegC complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
2OVP
 
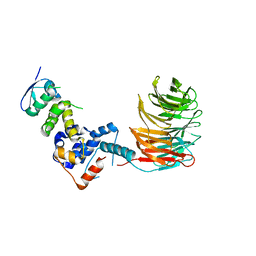 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
2L1E
 
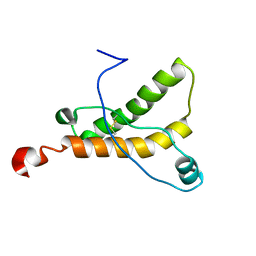 | | Mouse prion protein (121-231) containing the substitution F175A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2012-10-24 | | Method: | SOLUTION NMR | | Cite: | Prion Protein mPrP[F175A](121-231): Structure and Stability in Solution.
J.Mol.Biol., 423, 2012
|
|
2L1D
 
 | | Mouse prion protein (121-231) containing the substitution Y169G | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2011-11-09 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L1H
 
 | |
2L1K
 
 | | Mouse prion protein (121-231) containing the substitutions Y169A, Y225A, and Y226A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2012-02-01 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational exchange in the cellular form of prion proteins
To be Published
|
|
5BMF
 
 | | Crystal Structure of a Theophylline binding antibody Fab fragment | | Descriptor: | 2-(5-{1-[1-(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)-4,15-dioxo-8,11-dioxa-5,14-diazaicosan-20-yl]-3,3-dimethyl-6-sulfo-1,3-dihydro-2H-indol-2-ylidene}penta-1,3-dien-1-yl)-1-ethyl-3,3-dimethyl-3H-indolium-5-sulfonate, Fab fragment heavy chain, Fab fragment light chain | | Authors: | Bujotzek, A, Fuchs, A, Changtao, Q, Klostermann, S, Benz, J, Antes, I, Dengl, S, Hoffmann, E, Georges, G. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | MoFvAb: Modeling the Fv region of antibodies.
Mabs, 7, 2015
|
|
1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
6OY4
 
 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
1RI1
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
7QEN
 
 | | S.c. Condensin core in DNA- and ATP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Condensin complex subunit 1, Condensin complex subunit 2, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
7QFW
 
 | | S.c. Condensin peripheral Ycg1 subcomplex bound to DNA | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Synthetic DNA ligand, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
1RI5
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI2
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI4
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI3
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
6U5O
 
 | | Structure of the Human Metapneumovirus Polymerase bound to the phosphoprotein tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Pan, J, Qian, X, Lattmann, S, Sahili, A.E, Yeo, T.H, Kalocsay, M, Fearns, R, Lescar, J. | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the human metapneumovirus polymerase phosphoprotein complex.
Nature, 577, 2020
|
|
5CVS
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltoheptaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Rashid, A.M, Syson, K, Koliwer-Brandl, H, van de Weerd, R, Stevenson, C.E.M, Batey, S.F.D, Miah, F, Alber, M, Ioerger, T.R, Chandra, G, Appelmelk, B.J, Nartowski, K.P, Khimyak, Y.Z, Lawson, D.M, Jacobs, W.R, Geurtsen, J, Kalscheuer, R, Bornemann, S. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
1R3O
 
 | | Crystal structure of the first RNA duplex in L-conformation at 1.9A resolution | | Descriptor: | L-RNA | | Authors: | Vallazza, M, Perbandt, M, Klussmann, S, Rypniewski, W, Erdmann, V.A, Betzel, C. | | Deposit date: | 2003-10-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First look at RNA in L-configuration.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
5NPA
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
