3S41
 
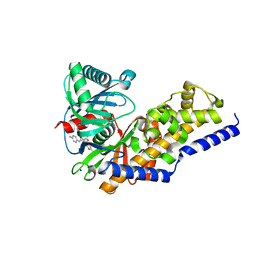 | | Glucokinase in complex with activator and glucose | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2011-05-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Designing glucokinase activators with reduced hypoglycemia risk: discovery of N,N-dimethyl-5-(2-methyl-6-((5-methylpyrazin-2-yl)-carbamoyl)benzofuran-4-yloxy)pyrimidine-2-carboxamide as a clinical candidate for the treatment of type 2 diabetes mellitus
MEDCHEMCOMM, 2, 2011
|
|
5DI1
 
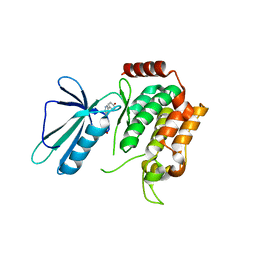 | | MAP4K4 in complex with an inhibitor | | Descriptor: | 4-{6-amino-5-[4-(methylsulfonyl)phenyl]pyridin-3-yl}phenol, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Liu, S. | | Deposit date: | 2015-08-31 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an in Vivo Tool to Establish Proof-of-Concept for MAP4K4-Based Antidiabetic Treatment.
Acs Med.Chem.Lett., 6, 2015
|
|
7EKI
 
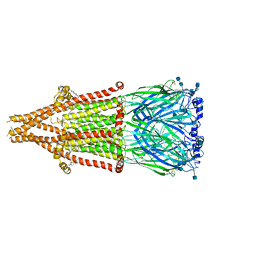 | | human alpha 7 nicotinic acetylcholine receptor in apo-form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKP
 
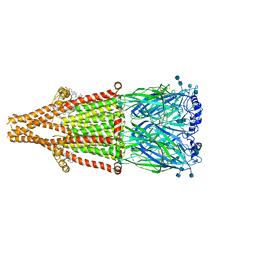 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKT
 
 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 and PNU-120596 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
6LA5
 
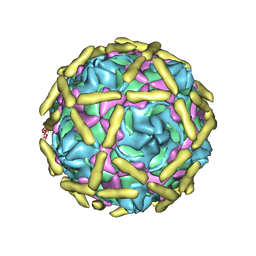 | |
6LB1
 
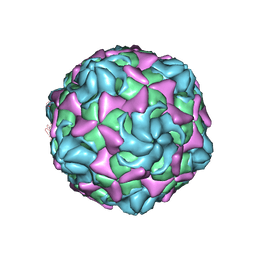 | | Cryo-EM structure of echovirus 11 A-particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA7
 
 | |
6LBO
 
 | | Cryo-EM structure of echovirus 11 empty particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA3
 
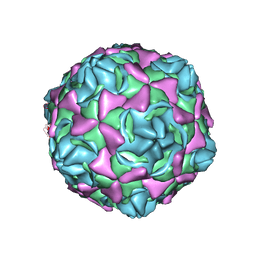 | | Cryo-EM structure of full echovirus 11 particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LAO
 
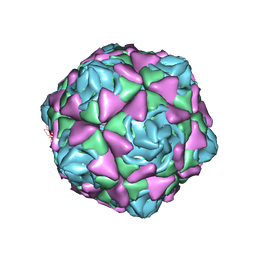 | |
6LBQ
 
 | | Cryo-EM structure of echovirus 11 empty particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
8CUH
 
 | | Crystal structure of human TEAD2 complexed with its inhibitor TM2. | | Descriptor: | 4-[3-(2-cyclohexylethoxy)benzoyl]-N-phenylpiperazine-1-carboxamide, Transcriptional enhancer factor TEF-4 | | Authors: | Liu, S, Luo, X. | | Deposit date: | 2022-05-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a new class of reversible TEA-domain transcription factor inhibitors with a novel binding mode.
Elife, 11, 2022
|
|
8FBD
 
 | | Crystal structure of OrfX1-OrfX3 complex from Clostridium botulinum E1 | | Descriptor: | ACETATE ION, Neurotoxin complex component Orf-X1, Neurotoxin complex component Orf-X3 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
8FBE
 
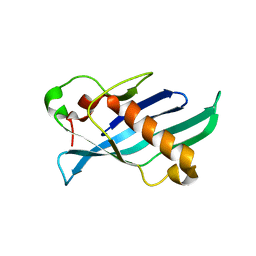 | | Crystal structure of OrfX1 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X1 | | Authors: | Liu, S, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
8JEU
 
 | |
8JET
 
 | |
8JEC
 
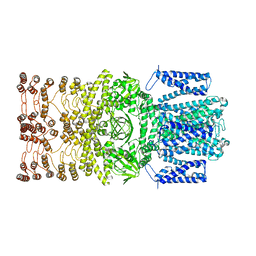 | |
6CDY
 
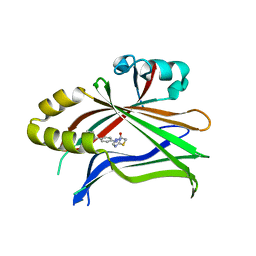 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
4NE9
 
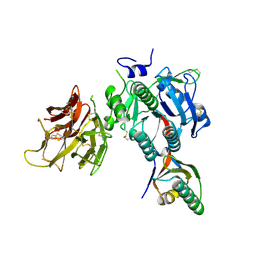 | | PCSK9 in complex with LDLR peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Liu, S. | | Deposit date: | 2013-10-28 | | Release date: | 2014-09-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and synthesis of truncated EGF-A peptides that restore LDL-R recycling in the presence of PCSK9 in vitro.
Chem.Biol., 21, 2014
|
|
7WBL
 
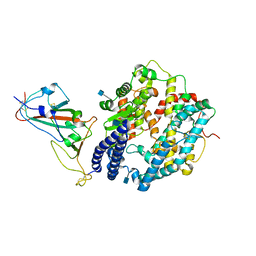 | | Cryo-EM structure of human ACE2 complexed with SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-01-19 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2.
Cell, 185, 2022
|
|
1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
