7P88
 
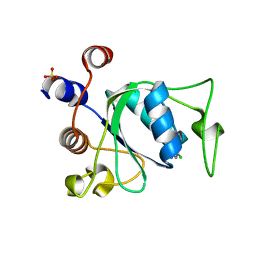 | | Crystal structure of YTHDC1 with compound YLI_DC1_002 | | Descriptor: | 2-chloranyl-~{N}-methyl-9~{H}-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8B
 
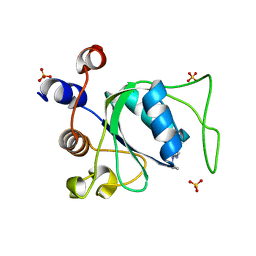 | | Crystal structure of YTHDC1 with compound YLI_DC1_006 | | Descriptor: | 9-cyclopropyl-~{N}-methyl-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8A
 
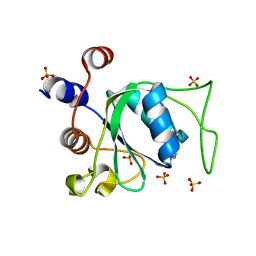 | | Crystal structure of YTHDC1 with compound YLI_DC1_003 | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N},9-dimethylpurin-6-amine | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P87
 
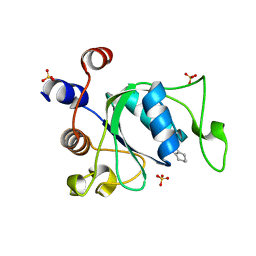 | | Crystal structure of YTHDC1 with compound YLI_DC1_001 | | Descriptor: | N-methyl-4,5,6,7-tetrahydro-1H-indazole-3-carboxamide, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8F
 
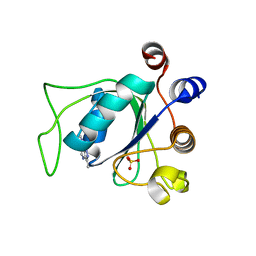 | | Crystal structure of YTHDC1 with compound YLI_DC1_008 | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N},3-dimethyl-1~{H}-pyrazolo[4,3-d]pyrimidin-7-amine | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
4N4G
 
 | |
4N4I
 
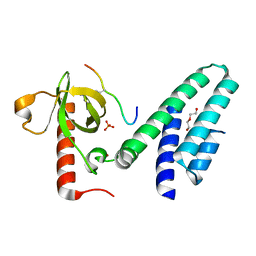 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4H
 
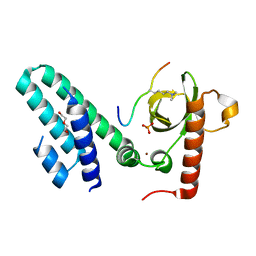 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
3DXF
 
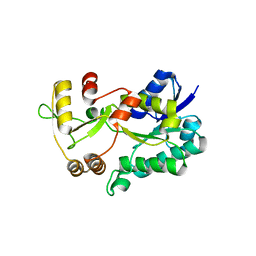 | | Crystal structure of the HSCARG R37A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-07-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
3E5M
 
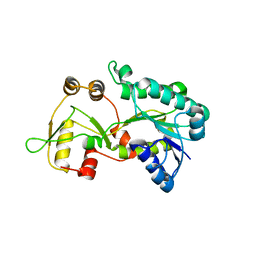 | | Crystal structure of the HSCARG Y81A mutant | | Descriptor: | NmrA-like family domain-containing protein 1 | | Authors: | Li, Y, Meng, G, Dai, X, Luo, M, Zheng, X. | | Deposit date: | 2008-08-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NADPH is an allosteric regulator of HSCARG
J.Mol.Biol., 387, 2009
|
|
1SMF
 
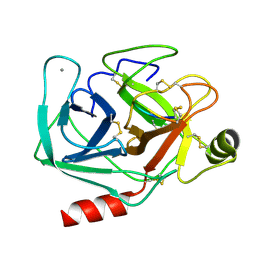 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
7F3B
 
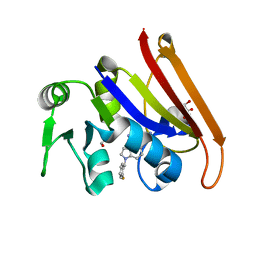 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
6ZD4
 
 | | Crystal structure of YTHDC1 S378A mutant | | Descriptor: | SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD3
 
 | | Crystal structure of YTHDC1 M438A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
8T5K
 
 | | Crystal structure of STING CTD in complex with BDW-OH | | Descriptor: | Stimulator of interferon genes protein, {[(4S)-8,9-dimethylthieno[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-3-yl]sulfanyl}acetic acid | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8T5L
 
 | | Crystal structure of STING CTD in complex with 2'3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
6ZD8
 
 | | Crystal structure of YTHDC1 T379V mutant | | Descriptor: | SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD5
 
 | |
6ZDA
 
 | |
6ZD7
 
 | |
4ZRK
 
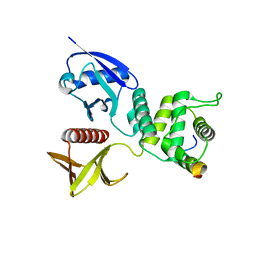 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
1NDM
 
 | |
1NDG
 
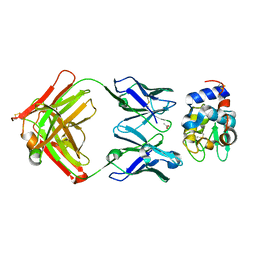 | | Crystal structure of Fab fragment of antibody HyHEL-8 complexed with its antigen lysozyme | | Descriptor: | ACETIC ACID, Lysozyme C, antibody kappa light chain, ... | | Authors: | Mariuzza, R.A, Li, Y, Li, H, Yang, F, Smith-Gill, S.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-06-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of the maturation of an antibody response to a protein antigen
Nat.Struct.Biol., 10, 2003
|
|
1FV1
 
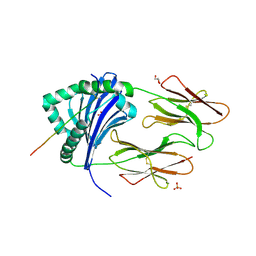 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
7R2E
 
 | | Structure of human Senp7 with SUMO2 | | Descriptor: | Sentrin-specific protease 7, Small ubiquitin-related modifier 3, prop-2-en-1-amine | | Authors: | Reverter, D, Li, Y. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis for the SUMO2 Isoform Specificity of SENP7.
J.Mol.Biol., 434, 2022
|
|
