4F4I
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form | | Descriptor: | Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form
To be Published
|
|
4I6V
 
 | | The crystal structure of an amidohydrolase 2 from Planctomyces limnophilus DSM 3776 | | Descriptor: | ACETATE ION, Amidohydrolase 2, GLYCEROL, ... | | Authors: | Fan, Y, Tan, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | The crystal structure of an amidohydrolase 2 from Planctomyces limnophilus DSM 3776
To be Published
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
4HYL
 
 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
4I66
 
 | |
4I6Z
 
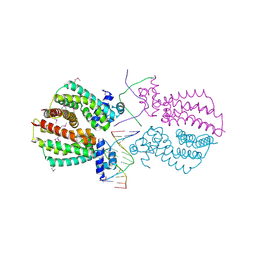 | | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide | | Descriptor: | DNA OLIGONUCLEOTIDE, Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide
To be Published
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4I76
 
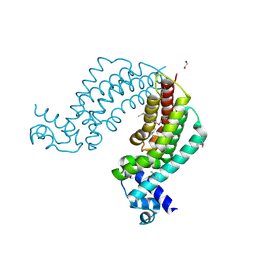 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
4MI1
 
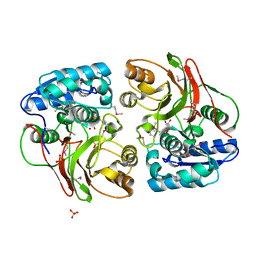 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Microcin immunity protein MccF, SULFATE ION | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
4M0C
 
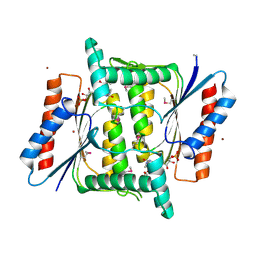 | | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN.
To be Published
|
|
4M0G
 
 | | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor. | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Tan, K, Zhou, M, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor.
To be Published
|
|
4MLZ
 
 | | Crystal structure of periplasmic binding protein from Jonesia denitrificans | | Descriptor: | CALCIUM ION, POTASSIUM ION, Periplasmic binding protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of periplasmic binding protein from Jonesia denitrificans
To be Published
|
|
4MJX
 
 | |
4MLC
 
 | | ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense | | Descriptor: | CALCIUM ION, Extracellular ligand-binding receptor, SULFATE ION | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense
To be Published
|
|
4MJM
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2544 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames
To be Published, 2013
|
|
4MK6
 
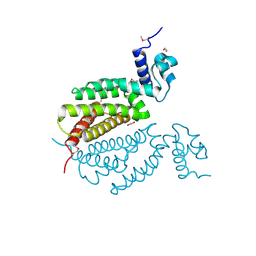 | | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, Probable Dihydroxyacetone Kinase Regulator DHSK_reg | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e
To be Published
|
|
4NHE
 
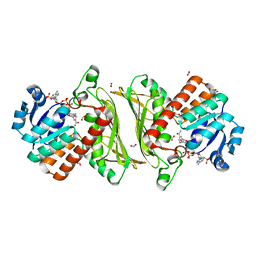 | | The crystal structure of oxidoreductase (Gfo/Idh/MocA family) from Streptococcus pneumoniae TIGR4 in complex with NADP | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-11-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of oxidoreductase (Gfo/Idh/MocA family) from Streptococcus pneumoniae TIGR4 in complex with NADP.
To be Published
|
|
4NEO
 
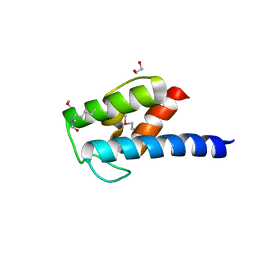 | | Structure of BlmI, a type-II acyl-carrier-protein from Streptomyces verticillus involved in bleomycin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Peptide synthetase NRPS type II-PCP | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Babnigg, G, Bruno, C.J.P, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins.
Proteins, 82, 2014
|
|
4NPB
 
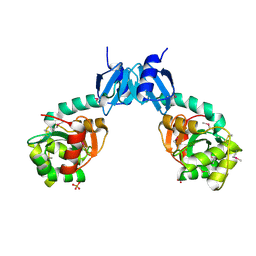 | | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92 | | Descriptor: | PHOSPHATE ION, Protein disulfide isomerase II, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92
To be Published
|
|
3O12
 
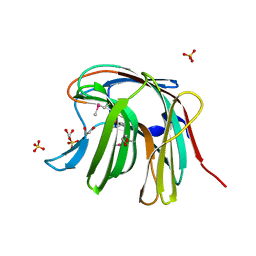 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
3O2I
 
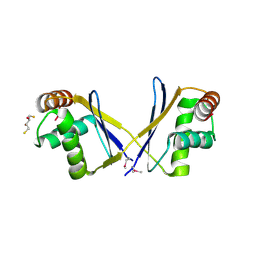 | | The crystal structure of a functionally unknown protein from Leptospirillum sp. Group II UBA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of a functionally unknown protein from Leptospirillum sp. Group II UBA
TO BE PUBLISHED
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4OVY
 
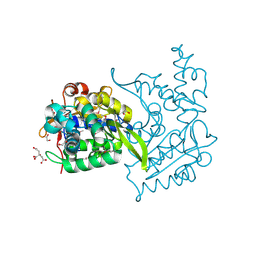 | | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Chang, C, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776
To be published
|
|
