7XWK
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWM
 
 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7X2U
 
 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | Authors: | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-02-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
1LTE
 
 | | STRUCTURE OF A LEGUME LECTIN WITH AN ORDERED N-LINKED CARBOHYDRATE IN COMPLEX WITH LACTOSE | | Descriptor: | CALCIUM ION, CORAL TREE LECTIN, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Lis, H, Sharon, N. | | Deposit date: | 1991-06-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a legume lectin with an ordered N-linked carbohydrate in complex with lactose.
Science, 254, 1991
|
|
7YRD
 
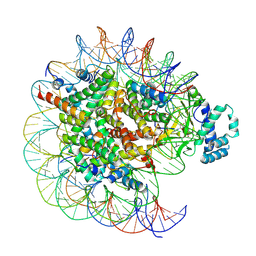 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7YRG
 
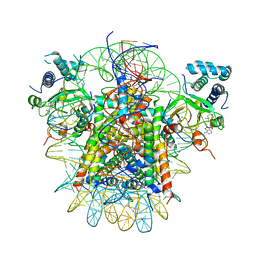 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
5ONU
 
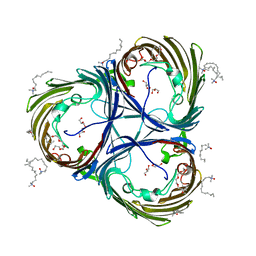 | | Trimeric OmpU structure | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Li, H.Y, Dong, C.J. | | Deposit date: | 2017-08-04 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the outer membrane protein OmpU from Vibrio cholerae at 2.2 angstrom resolution.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1FAG
 
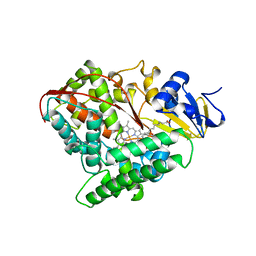 | | STRUCTURE OF CYTOCHROME P450 | | Descriptor: | CYTOCHROME P450 BM-3, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H.Y, Poulos, T.L. | | Deposit date: | 1996-08-01 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid.
Nat.Struct.Biol., 4, 1997
|
|
6LRM
 
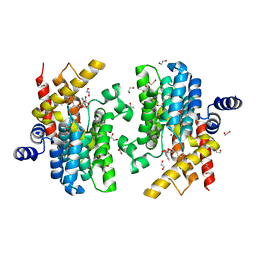 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
7CPU
 
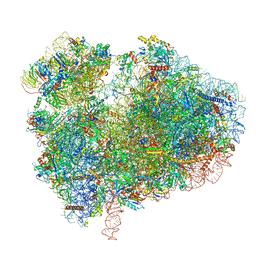 | | Cryo-EM structure of 80S ribosome from mouse kidney | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
1TVD
 
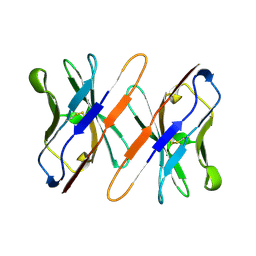 | | VARIABLE DOMAIN OF T CELL RECEPTOR DELTA CHAIN | | Descriptor: | T CELL RECEPTOR | | Authors: | Li, H.M, Lebedeva, M.I, Llera, A.S, Fields, B.A, Brenner, M.B, Mariuzza, R.A. | | Deposit date: | 1997-10-19 | | Release date: | 1998-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Vdelta domain of a human gammadelta T-cell antigen receptor.
Nature, 391, 1998
|
|
1AC6
 
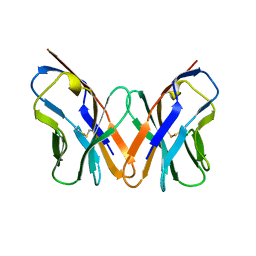 | |
2H80
 
 | | NMR structures of SAM domain of Deleted in Liver Cancer 2 (DLC2) | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Li, H.Y, Fung, K.L, Jin, D.Y, Chung, S.S, Ko, B.C, Sun, H.Z. | | Deposit date: | 2006-06-06 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Proteins, 67, 2007
|
|
7N1R
 
 | | A novel and unique ATP hydrolysis to AMP by a human Hsp70 BiP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Yang, J, Musayev, F, Liu, Q. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel and unique ATP hydrolysis to AMP by a human Hsp70 Binding immunoglobin protein (BiP).
Protein Sci., 31, 2022
|
|
3FRZ
 
 | | Crystal Structure of HCV NS5B RNA polymerase in complex with PF868554 | | Descriptor: | (6R)-6-cyclopentyl-6-[2-(2,6-diethylpyridin-4-yl)ethyl]-3-[(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl]-4-hydroxy-5,6-dihydro-2H-pyran-2-one, BETA-MERCAPTOETHANOL, N-[(benzyloxy)carbonyl]-L-alpha-glutamyl-N-[(1S)-4-oxo-4-phenyl-1-propylbut-2-en-1-yl]-L-phenylalaninamide, ... | | Authors: | Parge, H.E. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor.
J.Med.Chem., 52, 2009
|
|
1NOO
 
 | | CYTOCHROME P450-CAM COMPLEXED WITH 5-EXO-HYDROXYCAMPHOR | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H.Y, Poulos, T.L. | | Deposit date: | 1995-12-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Cytochrome P450-Cam Complexed with its Catalytic Product, 5-Exo-Hydroxycamphor
J.Am.Chem.Soc., 117, 1995
|
|
2FUI
 
 | |
2FUU
 
 | |
8GTX
 
 | |
7XVI
 
 | |
7XVK
 
 | |
7N8C
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-06-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
6XRT
 
 | |
