8CW7
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW6
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW3
 
 | | 20us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWB
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWE
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVV
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWD
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVW
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWF
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
5TTC
 
 | | XFEL structure of influenza A M2 wild type TM domain at high pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-11-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UM1
 
 | | XFEL structure of influenza A M2 wild type TM domain at intermediate pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CCI
 
 | | Structure of the Mg2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | MAGNESIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5CCH
 
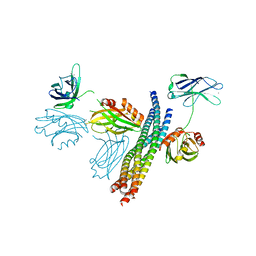 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5CCJ
 
 | |
5CCG
 
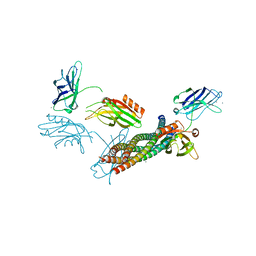 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5F66
 
 | |
5F6M
 
 | | Isotropic Trypsin Model for Comparison of Diffuse Scattering | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Van Benschoten, A.H, Wall, M.E, Fraser, J.S. | | Deposit date: | 2015-12-06 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Measuring and modeling diffuse scattering in protein X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6H6K
 
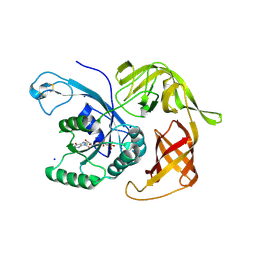 | | The structure of the FKR mutant of the archaeal translation initiation factor 2 gamma subunit in complex with GDPCP, obtained in the absence of magnesium salts in the crystallization solution. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, SODIUM ION, ... | | Authors: | Nikonov, O, Kravchenko, O, Nevskaya, N, Stolboushkina, E, Gabdulkhakov, A, Garber, M, Nikonov, S. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7Z3E
 
 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with hydroquinone NrdI from Bacillus cereus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
7Z3D
 
 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with oxidized NrdI from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
6YDI
 
 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, diferrous state | | Descriptor: | FE (II) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
6Y0P
 
 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | Descriptor: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
