5Q0J
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-[2-(5-phenylthiophen-2-yl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0X
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q15
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q11
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,N-dicyclohexyl-3-(2,4-dichlorophenyl)-5-methyl-1,2-oxazole-4-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q18
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
1POZ
 
 | | SOLUTION STRUCTURE OF THE HYALURONAN BINDING DOMAIN OF HUMAN CD44 | | Descriptor: | CD44 antigen | | Authors: | Teriete, P, Banerji, S, Blundell, C.D, Kahmann, J.D, Pickford, A.R, Wright, A.J, Campbell, I.D, Jackson, D.G, Day, A.J. | | Deposit date: | 2003-06-16 | | Release date: | 2004-03-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44.
Mol.Cell, 13, 2004
|
|
5VKI
 
 | | Crystal structure of P[19] rotavirus VP8* complexed with mucin core 2 | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
5VKS
 
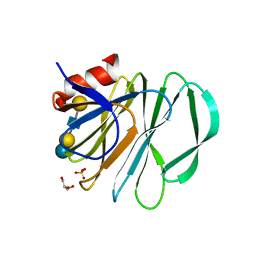 | | Crystal structure of P[19] rotavirus VP8* complexed with LNFPI | | Descriptor: | GLYCEROL, Outer capsid protein VP4, SULFATE ION, ... | | Authors: | Xu, S, Liu, Y, Woodruff, A, Zhong, W, Jiang, X, Kennedy, M.A. | | Deposit date: | 2017-04-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of glycan specificity of P[19] VP8*: Implications for rotavirus zoonosis and evolution.
PLoS Pathog., 13, 2017
|
|
4EYW
 
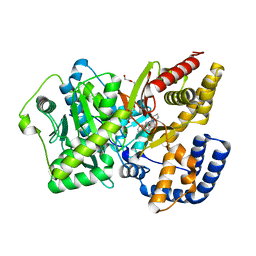 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with 1-[(R)-2-(3,4-Dihydro-1H-isoquinoline-2-carbonyl)-piperidin-1-yl]-2-phenoxy-ethanone | | Descriptor: | 1-[(2R)-2-(3,4-dihydroisoquinolin-2(1H)-ylcarbonyl)piperidin-1-yl]-2-phenoxyethanone, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Rufer, A, Joseph, C. | | Deposit date: | 2012-05-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
8B30
 
 | |
7ZW8
 
 | |
7ZY6
 
 | |
5BSA
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BS7
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
3QUM
 
 | | Crystal structure of human prostate specific antigen (PSA) in Fab sandwich with a high affinity and a PCa selective antibody | | Descriptor: | Fab 5D3D11 Heavy Chain, Fab 5D3D11 Light Chain, Fab 5D5A5 Heavy Chain, ... | | Authors: | Stura, E.A, Muller, B.H, Michel, S, Ducancel, F. | | Deposit date: | 2011-02-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostate-specific antigen in a sandwich antibody complex.
J.Mol.Biol., 414, 2011
|
|
8AEP
 
 | | Reductase domain of the carboxylate reductase of Neurospora crassa | | Descriptor: | Acetyl-CoA synthetase-like protein, CHLORIDE ION, SULFATE ION | | Authors: | Daniel, B, Schrufer, A, Marlene, L, Sagmeister, T, Pavkov-Keller, T. | | Deposit date: | 2022-07-13 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Reductase Domain of a Fungal Carboxylic Acid Reductase and Its Substrate Scope in Thioester and Aldehyde Reduction.
Acs Catalysis, 12, 2022
|
|
