7OEZ
 
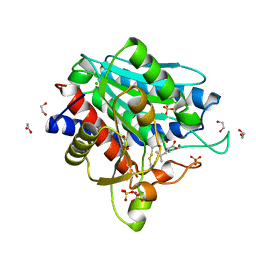 | | Leucine Aminopeptidase A mature enzyme in a complex with leucine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Watson, K.A, Baltulionis, G. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The role of propeptide-mediated autoinhibition and intermolecular chaperone in the maturation of cognate catalytic domain in leucine aminopeptidase.
J.Struct.Biol., 213, 2021
|
|
3I3T
 
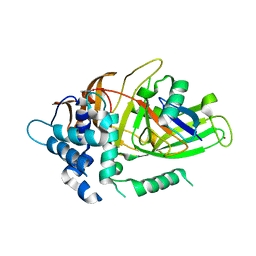 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
5DZ8
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA variable (V) domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BspA (BspA_V), ... | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
6CPI
 
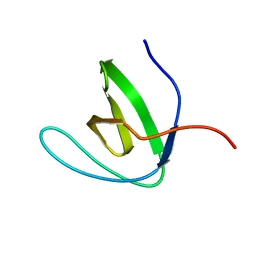 | | Solution structure of SH3 domain from Shank1 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPK
 
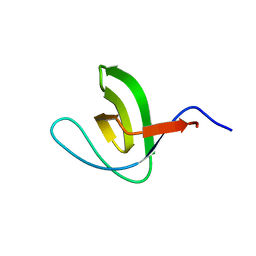 | | Solution structure of SH3 domain from Shank3 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPJ
 
 | | Solution structure of SH3 domain from Shank2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 2 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
5DZ9
 
 | |
5DZA
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA C terminal domain (WT) | | Descriptor: | 1,2-ETHANEDIOL, BspA, DI(HYDROXYETHYL)ETHER | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
5LBT
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with imiquimod | | Descriptor: | 1-(2-methylpropyl)imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
5LBU
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with to CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
6F35
 
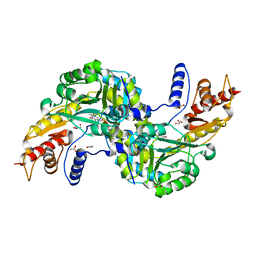 | | Crystal structure of the aspartate aminotranferase from Rhizobium meliloti | | Descriptor: | ACETATE ION, Aspartate aminotransferase B, GLYCEROL, ... | | Authors: | Cobessi, D, Graindorge, M, Giustini, C, Matringe, M. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6F77
 
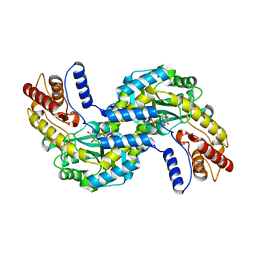 | | Crystal structure of the prephenate aminotransferase from Rhizobium meliloti | | Descriptor: | Aspartate aminotransferase A, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cobessi, D, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
3HRF
 
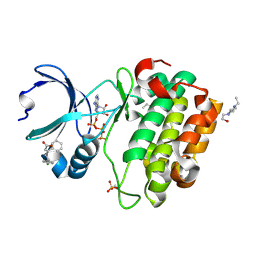 | | Crystal structure of Human PDK1 kinase domain in complex with an allosteric activator bound to the PIF-pocket | | Descriptor: | (2Z)-5-(4-chlorophenyl)-3-phenylpent-2-enoic acid, 3-phosphoinositide-dependent protein kinase 1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hindie, V, Alzari, P.M, Biondi, R.M. | | Deposit date: | 2009-06-09 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1.
Nat.Chem.Biol., 5, 2009
|
|
6G4G
 
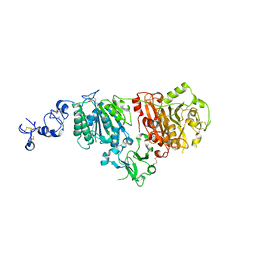 | | Full length ectodomain of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) including the SMB domains but with a partially disordered active site structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dohler, C, Zebisch, M, Strater, N. | | Deposit date: | 2018-03-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallization of ectonucleotide phosphodiesterase/pyrophosphatase-3 and orientation of the SMB domains in the full-length ectodomain.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3U3A
 
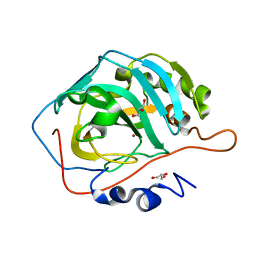 | |
5TFT
 
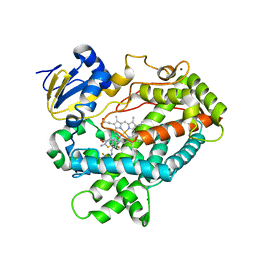 | | Structure of cytochrome P450 2D6 (CYP2D6) BACE1 inhibitor complex | | Descriptor: | (4S)-4-[2,4-difluoro-5-({[1-(trifluoromethyl)cyclopropyl]amino}methyl)phenyl]-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2016-09-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality.
J. Med. Chem., 60, 2017
|
|
1J6U
 
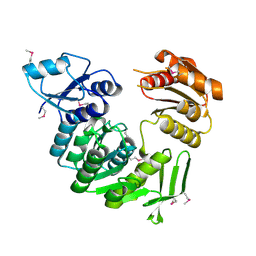 | |
1HE7
 
 | | Human Nerve growth factor receptor TrkA | | Descriptor: | GLYCEROL, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Banfield, M, Robertson, A, Allen, S, Dando, J, Tyler, S, Bennett, G, Brain, S, Mason, G, Holden, P, Clarke, A, Naylor, R, Wilcock, G, Brady, R, Dawbarn, D. | | Deposit date: | 2000-11-20 | | Release date: | 2001-04-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Structure of the Nerve Growth Factor Binding Site on Trka.
Biochem.Biophys.Res.Commun., 282, 2001
|
|
5TFU
 
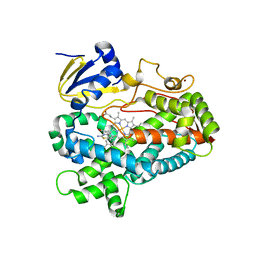 | | Structure of cytochrome P450 2D6 (CYP2D6) BACE1 inhibitor complex | | Descriptor: | (4S,6R)-4-[2,4-difluoro-5-({[1-(trifluoromethyl)cyclopropyl]amino}methyl)phenyl]-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2016-09-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality.
J. Med. Chem., 60, 2017
|
|
3U47
 
 | | Human Carbonic Anhydrase II V143L | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | West, D, Mckenna, R. | | Deposit date: | 2011-10-07 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural modification of the hydrophobic pocket of Human Carbonic Anhydrase II
To be Published
|
|
5T1W
 
 | | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluoro-5-{[(2,2,2-trifluoroethyl)amino]methyl}phenyl)-6-(fluoromethyl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, CHLORIDE ION, ... | | Authors: | Parris, K.D, Vajdos, F. | | Deposit date: | 2016-08-22 | | Release date: | 2017-01-11 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality.
J. Med. Chem., 60, 2017
|
|
5T1U
 
 | | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality | | Descriptor: | (4S)-4-[2,4-difluoro-5-({[1-(trifluoromethyl)cyclopropyl]amino}methyl)phenyl]-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Parris, K.D, Vajdos, F. | | Deposit date: | 2016-08-22 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Aminomethyl-Derived Beta Secretase (BACE1) Inhibitors: Engaging Gly230 without an Anilide Functionality.
J. Med. Chem., 60, 2017
|
|
3U45
 
 | | Human Carbonic Anhydrase II V143A | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | West, D, Mckenna, R. | | Deposit date: | 2011-10-07 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural Modification of the hydrophobic pocket in Human Carbonic Anhydrase II
To be Published
|
|
1J5S
 
 | |
4FCB
 
 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 3,4-dimethyl-1-propyl-7-(quinolin-2-ylmethoxy)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
