6LLY
 
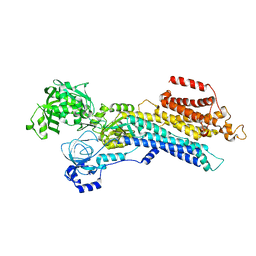 | | CryoEM structure of SERCA2b WT in E2-BeF3- state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
3UG9
 
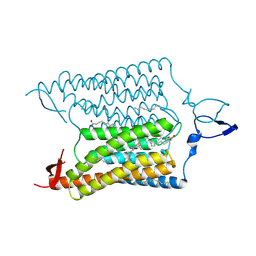 | | Crystal Structure of the Closed State of Channelrhodopsin | | Descriptor: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | Authors: | Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2011-11-02 | | Release date: | 2012-01-25 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
5H2O
 
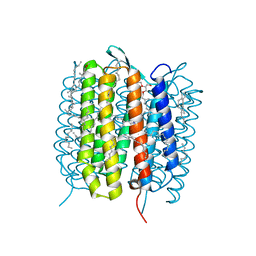 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2L
 
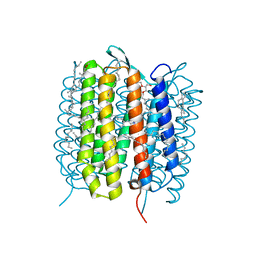 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 5.25 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2M
 
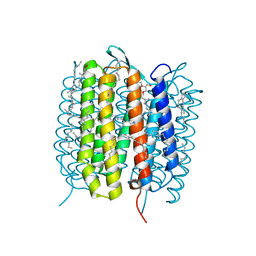 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 13.8 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2P
 
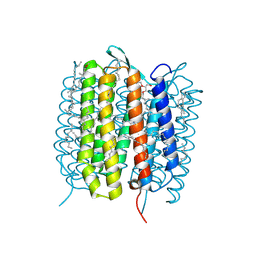 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2J
 
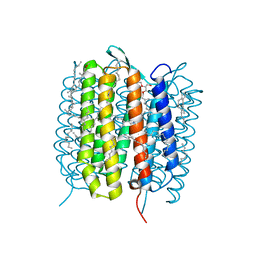 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2H
 
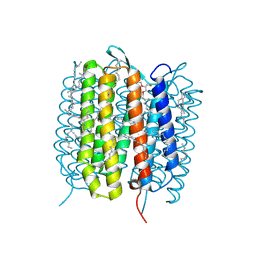 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 40 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2K
 
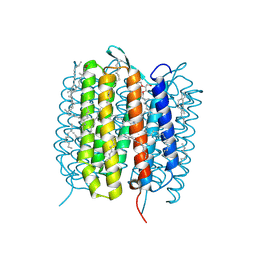 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2I
 
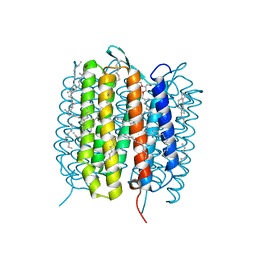 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 110 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2N
 
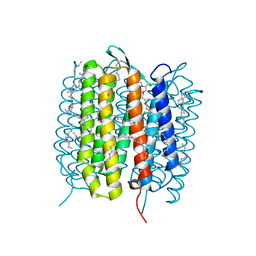 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 95.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5ZSU
 
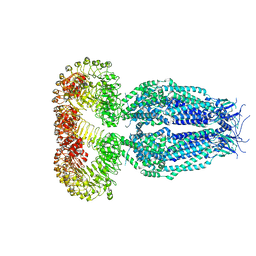 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8IJM
 
 | | Cyo-EM structure of K794A non-gastric proton pump in Na+ bound E1AMPPCP state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | An unusual conformation from Na + -sensitive non-gastric proton pump mutants reveals molecular mechanisms of cooperative Na + -binding.
Biochim Biophys Acta Mol Cell Res, 1870, 2023
|
|
8IJL
 
 | | Cyo-EM structure of wildtype non-gastric proton pump in the presence of Na+, AlF and ADP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Abe, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | An unusual conformation from Na + -sensitive non-gastric proton pump mutants reveals molecular mechanisms of cooperative Na + -binding.
Biochim Biophys Acta Mol Cell Res, 1870, 2023
|
|
5I20
 
 | | Crystal structure of protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | Authors: | Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
6LN5
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN9
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class2) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN7
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class3) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN8
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class1) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN6
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class2) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
4GEL
 
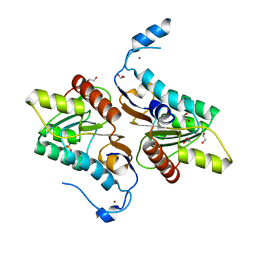 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEN
 
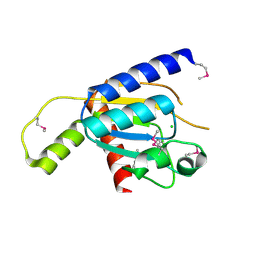 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
7VTN
 
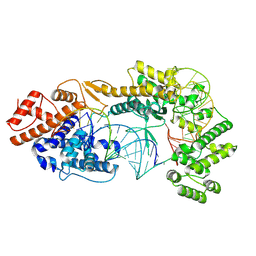 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
4GEM
 
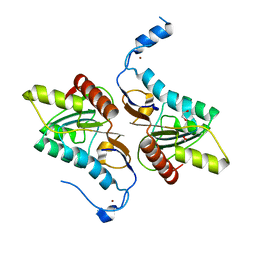 | | Crystal structure of Zucchini (K171A) | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
7CYN
 
 | | Cryo-EM structure of human TLR7 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
