3F38
 
 | | Apoferritin: complex with 2,6-dimethylphenol | | Descriptor: | 2,6-dimethylphenol, CADMIUM ION, Ferritin light chain, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
3F32
 
 | | Horse spleen apoferritin | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
3F37
 
 | | Apoferritin: complex with 2,6-dimethylphenol | | Descriptor: | 2,6-dimethylphenol, ACETATE ION, CADMIUM ION, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
9H1Z
 
 | |
3F35
 
 | | Apoferritin: complex with 2,6-diethylphenol | | Descriptor: | 2,6-diethylphenol, CADMIUM ION, Ferritin light chain, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
3F33
 
 | | Apoferritin: complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
3F34
 
 | | Apoferritin: complex with 2,6-diethylphenol | | Descriptor: | 2,6-diethylphenol, ACETATE ION, CADMIUM ION, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
2GYD
 
 | | Complex of equine apoferritin with the H-diaziflurane photolabeling reagent | | Descriptor: | 2-(1,1-DIFLUOROETHOXY)-1,1,1-TRIFLUOROETHANE, CADMIUM ION, ferritin L subunit | | Authors: | Loll, P.J, Rossi, M.J, Eckenhoff, R. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Photoactive analogues of the haloether anesthetics provide high-resolution features from low-affinity interactions.
Acs Chem.Biol., 1, 2006
|
|
8UZH
 
 | |
3LA1
 
 | |
6JC4
 
 | |
7D3Y
 
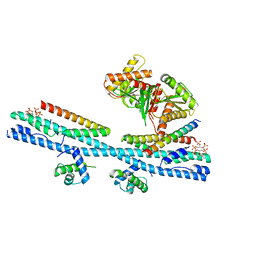 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
7D3T
 
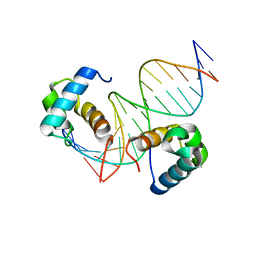 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
6L94
 
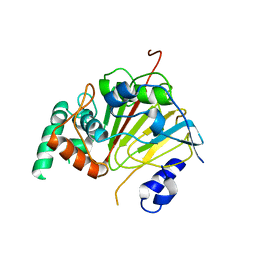 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
5DV4
 
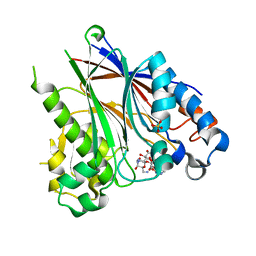 | |
5EFW
 
 | | Crystal structure of LOV2-Zdk1 - the complex of oat LOV2 and the affibody protein Zdark1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NPH1-1, SULFATE ION, ... | | Authors: | Winkler, A, Wang, H, Hartmann, E, Hahn, K, Schlichting, I. | | Deposit date: | 2015-10-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
5DJU
 
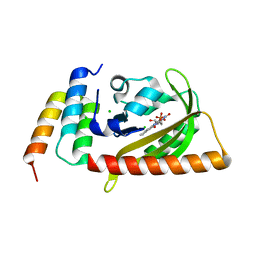 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk3 | | Descriptor: | CHLORIDE ION, Engineered protein, Zdk3 affibody, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
5DV2
 
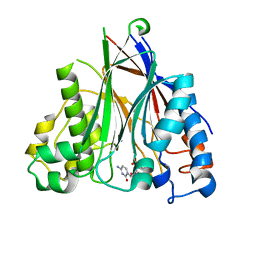 | |
5DJT
 
 | | Crystal structure of LOV2 (C450A) domain in complex with Zdk2 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Engineered protein, ... | | Authors: | Tarnawski, M, Wang, H, Yumerefendi, H, Hahn, K.M, Schlichting, I. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
4ZHU
 
 | | Crystal structure of a bacterial repressor protein | | Descriptor: | SULFATE ION, YfiR | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3968 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHW
 
 | |
4ZHY
 
 | | Crystal structure of a bacterial signalling complex | | Descriptor: | FORMIC ACID, SULFATE ION, YfiB, ... | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHV
 
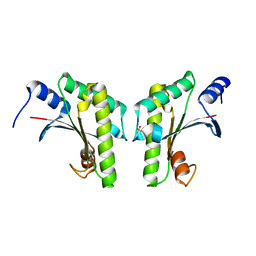 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
7CVQ
 
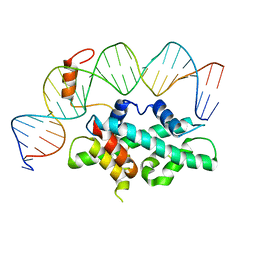 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
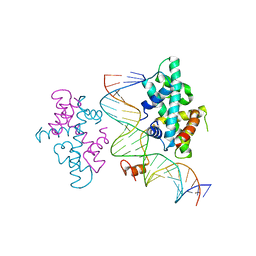 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
