1NNY
 
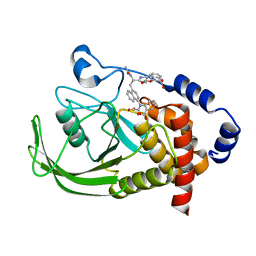 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 23 Using a Linked-Fragment Strategy | | Descriptor: | 3-({5-[(N-ACETYL-3-{4-[(CARBOXYCARBONYL)(2-CARBOXYPHENYL)AMINO]-1-NAPHTHYL}-L-ALANYL)AMINO]PENTYL}OXY)-2-NAPHTHOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1Q59
 
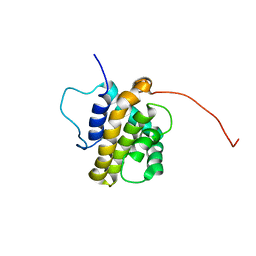 | | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2 | | Descriptor: | Early antigen protein R | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2
J.Mol.Biol., 332, 2003
|
|
1DHM
 
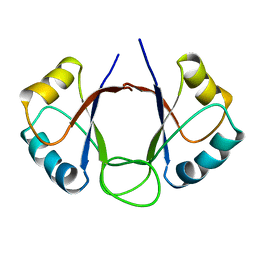 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
1GJH
 
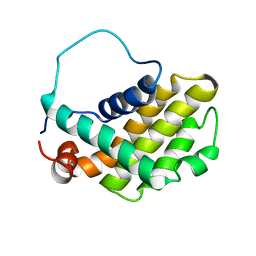 | | HUMAN BCL-2, ISOFORM 2 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G5J
 
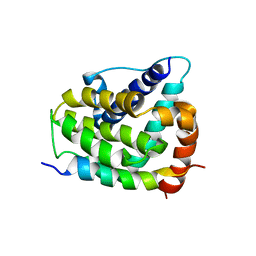 | | COMPLEX OF BCL-XL WITH PEPTIDE FROM BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BAD PROTEIN | | Authors: | Petros, A.M, Nettesheim, D.G, Wang, Y, Olejniczak, E.T, Meadows, R.P, Mack, J, Swift, K, Matayoshi, E.D, Zhang, H, Thompson, C.B, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies.
Protein Sci., 9, 2000
|
|
1G5M
 
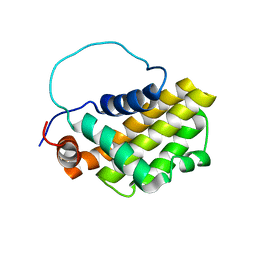 | | HUMAN BCL-2, ISOFORM 1 | | Descriptor: | PROTEIN (APOPTOSIS REGULATOR BCL-2 WITH PUTATIVE FLEXIBLE LOOP REPLACED WITH A PORTION OF APOPTOSIS REGULATOR BCL-X PROTEIN) | | Authors: | Petros, A.M, Medek, A, Nettesheim, D.G, Kim, D.H, Yoon, H.S, Swift, K, Matayoshi, E.D, Oltersdorf, T, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antiapoptotic protein bcl-2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G3F
 
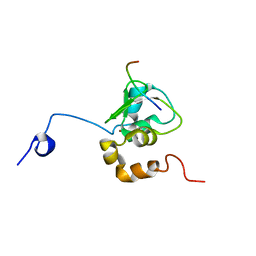 | | NMR STRUCTURE OF A 9 RESIDUE PEPTIDE FROM SMAC/DIABLO COMPLEXED TO THE BIR3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN 3, SMAC, ZINC ION | | Authors: | Liu, Z, Sun, C, Olejniczak, E.T, Meadows, R.P, Betz, S.F, Oost, T, Herrmann, J, Wu, J.C, Fesik, S.W. | | Deposit date: | 2000-10-24 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain.
Nature, 408, 2000
|
|
1F9X
 
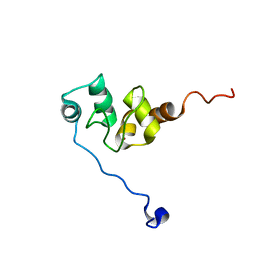 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | Authors: | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | Deposit date: | 2000-07-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
5UJB
 
 | | Structure of a Mcl-1 Inhibitor Binding to Site 3 of Human Serum Albumin | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, PHOSPHATE ION, Serum albumin | | Authors: | Zhao, B. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Myeloid cell leukemia-1 (Mcl-1) inhibitor bound to drug site 3 of Human Serum Albumin.
Bioorg. Med. Chem., 25, 2017
|
|
2QFO
 
 | | HSP90 complexed with A143571 and A516383 | | Descriptor: | (3E)-3-[(phenylamino)methylidene]dihydrofuran-2(3H)-one, 4-METHYL-6-(TRIFLUOROMETHYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
CHEM.BIOL.DRUG DES., 70, 2007
|
|
1IRS
 
 | | IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IL-4 RECEPTOR PHOSPHOPEPTIDE, IRS-1 | | Authors: | Zhou, M.-M, Huang, B, Olejniczak, E.T, Meadows, R.P, Shuker, S.B, Miyazaki, M, Trub, T, Shoelson, S.E, Feisk, S.W. | | Deposit date: | 1996-03-22 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain.
Nat.Struct.Biol., 3, 1996
|
|
4ZBF
 
 | | Mcl-1 complexed with small molecules | | Descriptor: | (1R)-7-[3-(naphthalen-1-yloxy)propyl]-3,4-dihydro-2H-[1,4]thiazepino[2,3,4-hi]indole-6-carboxylic acid 1-oxide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
7ACF
 
 | | CRYSTAL STRUCTURE OF CRYSTAL FORM 2 OF AN ACTIVE KRAS G12D (GPPCP) DIMER IN COMPLEX WITH BI-5747 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[6-[[3-[(1~{S})-6-oxidanyl-3-oxidanylidene-1,2-dihydroisoindol-1-yl]-1~{H}-indol-2-yl]methylamino]hexylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Reply to Tran et al.: Dimeric KRAS protein-protein interaction stabilizers.
Proc. Natl. Acad. Sci. U.S.A., 117, 2020
|
|
7U8H
 
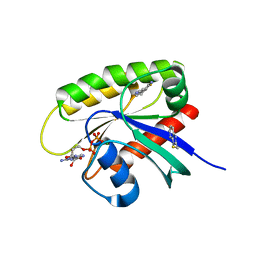 | | Discovery of a KRAS G12V Inhibitor in vivo Tool Compound starting from an HSQC-NMR based Fragment Hit | | Descriptor: | 1H-benzimidazol-2-ylmethanethiol, 2-amino-4,5,6,7-tetrahydro-1-benzothiophene-3-carbonitrile, GTPase KRas, ... | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor.
J.Med.Chem., 65, 2022
|
|
4ZBI
 
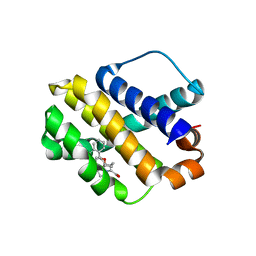 | | Mcl-1 complexed with small molecules | | Descriptor: | 1-[3-(naphthalen-1-yloxy)propyl]-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
7U9Y
 
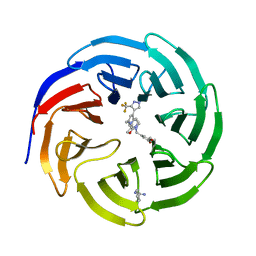 | |
7UAS
 
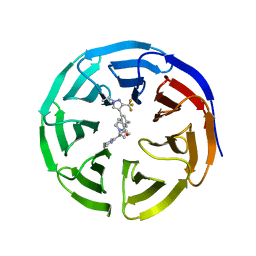 | |
5IF4
 
 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
5IEZ
 
 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 3-({6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-1H-indole-2-carbonyl}amino)benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
6GJ6
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 18 | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A, Phan, J. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GJ8
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH BI 2852 | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1TCE
 
 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2QF6
 
 | | HSP90 complexed with A56322 | | Descriptor: | 6-(3-BROMO-2-NAPHTHYL)-1,3,5-TRIAZINE-2,4-DIAMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG2
 
 | | HSP90 complexed with A917985 | | Descriptor: | 3-({2-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)ETHYNYL]BENZYL}AMINO)-1,3-OXAZOL-2(3H)-ONE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG0
 
 | | HSP90 complexed with A943037 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)METHYL]-3-{[(E)-(2-OXODIHYDROFURAN-3(2H)-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
