3KL1
 
 | |
5HWL
 
 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | Descriptor: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Zhang, X, Zhang, L, Zhang, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-02-13 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (6.78 Å) | | Cite: | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
3IYL
 
 | | Atomic CryoEM Structure of a Nonenveloped Virus Suggests How Membrane Penetration Protein is Primed for Cell Entry | | Descriptor: | Core protein VP6, MYRISTIC ACID, Outer capsid VP4, ... | | Authors: | Zhang, X, Jin, L, Fang, Q, Hui, W, Zhou, Z.H. | | Deposit date: | 2010-02-02 | | Release date: | 2010-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry.
Cell(Cambridge,Mass.), 141, 2010
|
|
4DXW
 
 | | Crystal structure of NavRh, a voltage-gated sodium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Zhang, X, Ren, W.L, Yan, C.Y, Wang, J.W, Yan, N. | | Deposit date: | 2012-02-28 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel
Nature, 486, 2012
|
|
3J4U
 
 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | Descriptor: | cementing protein, major capsid protein | | Authors: | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
2GS6
 
 | | Crystal Structure of the active EGFR kinase domain in complex with an ATP analog-peptide conjugate | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, Peptide, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2010-09-04 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
2GS7
 
 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS2
 
 | | Crystal Structure of the active EGFR kinase domain | | Descriptor: | Epidermal growth factor receptor | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor.
Cell(Cambridge,Mass.), 125, 2006
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
1F3L
 
 | |
2RF9
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4GJT
 
 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
5K3H
 
 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
5K3I
 
 | | Crystal structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans complexed with FAD and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1H0C
 
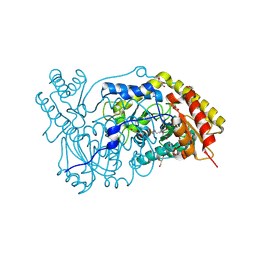 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
5K3G
 
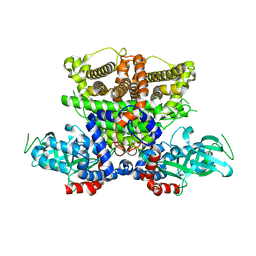 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2RFD
 
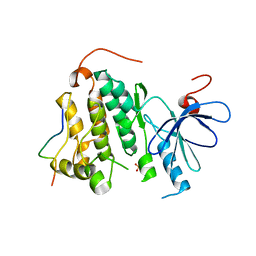 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
