5P4O
 
 | |
5P87
 
 | |
5P53
 
 | |
5P8M
 
 | |
5P5G
 
 | |
5P5X
 
 | |
5P6C
 
 | |
5P6Q
 
 | |
5P76
 
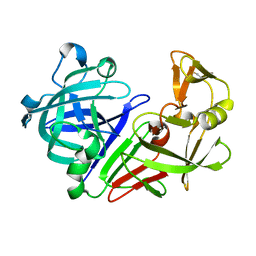 | |
1PPQ
 
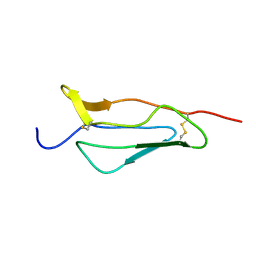 | | NMR structure of 16th module of Immune Adherence Receptor, Cr1 (Cd35) | | Descriptor: | Complement receptor type 1 | | Authors: | O'Leary, J.M, Bromek, K, Black, G.M, Uhrinova, S, Schmitz, C, Krych, M, Atkinson, J.P, Uhrin, D, Barlow, P.N. | | Deposit date: | 2003-06-17 | | Release date: | 2004-05-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces.
Protein Sci., 13, 2004
|
|
1M8S
 
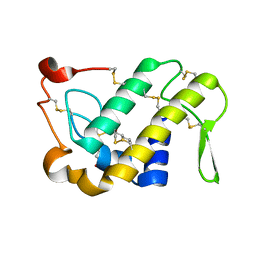 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1M8R
 
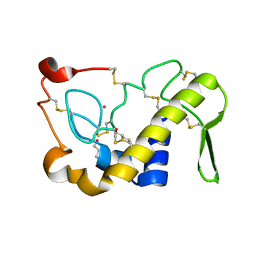 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
3GXA
 
 | | Crystal structure of GNA1946 | | Descriptor: | METHIONINE, Outer membrane lipoprotein GNA1946, SULFATE ION | | Authors: | Yang, X, Shen, Y. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of lipoprotein GNA1946 from Neisseria meningitidis
J.Struct.Biol., 168, 2009
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5HQA
 
 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
3L9N
 
 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5HQB
 
 | | A Glycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
3L9L
 
 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5HQ4
 
 | | A Glycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
7M5Z
 
 | | Crystal Structure of the MerTK Kinase Domain in Complex with Inhibitor MIPS15692 | | Descriptor: | 2-(butylamino)-N-[1-(3-fluoropropyl)piperidin-4-yl]-4-{[(1r,4r)-4-hydroxycyclohexyl]amino}pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Hermans, S.J, Hancock, N.C, Baell, J.B, Parker, M.W. | | Deposit date: | 2021-03-25 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Development of [ 18 F]MIPS15692, a radiotracer with in vitro proof-of-concept for the imaging of MER tyrosine kinase (MERTK) in neuroinflammatory disease.
Eur.J.Med.Chem., 226, 2021
|
|
6X9T
 
 | |
8CSG
 
 | | Human PRMT5:MEP50 structure with Fragment 1 and MTA Bound | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-bromo-1H-pyrrolo[3,2-b]pyridin-5-amine, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7MT4
 
 | | Crystal structure of tryptophan Synthase in complex with F9, NH4+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMMONIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XG4
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase L28R mutant in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
6XG5
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
