2RR2
 
 | | Structure of O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1, alpha-L-fucopyranose | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
1WP8
 
 | | crystal structure of Hendra Virus fusion core | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Nipah and Hendra virus fusion core proteins
FEBS J., 273, 2006
|
|
7NOX
 
 | | Structure of SGBP BO2743 from Bacteroides ovatus in complex with mixed-linked gluco-nonasaccharide | | Descriptor: | AZIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Correia, V.C, Trovao, F, Pinheiro, B.A, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mapping Molecular Recognition of beta 1,3-1,4-Glucans by a Surface Glycan-Binding Protein from the Human Gut Symbiont Bacteroides ovatus.
Microbiol Spectr, 9, 2021
|
|
7EKQ
 
 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7TXI
 
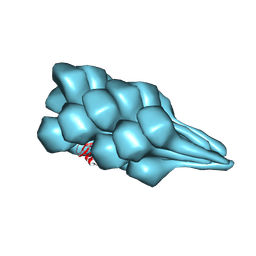 | | Cryo-EM of A. pernix flagellum | | Descriptor: | Probable flagellin 1 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2RR0
 
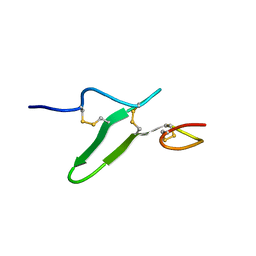 | | Structure of epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
7MDN
 
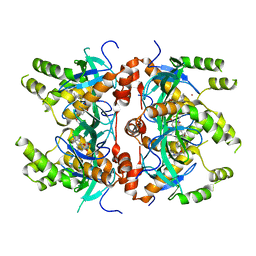 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
2RQZ
 
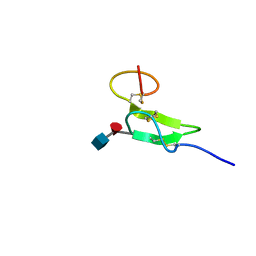 | | Structure of sugar modified epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, Neurogenic locus notch homolog protein 1 | | Authors: | Shimizu, K, Fujitani, N, Hosoguchi, K, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2PG8
 
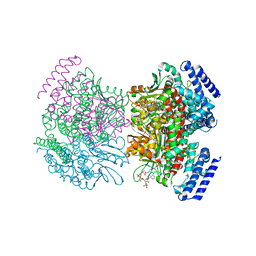 | | Crystal structure of R254K mutanat of DpgC with bound substrate analog | | Descriptor: | DpgC, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Fielding, E.N. | | Deposit date: | 2007-04-09 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate Recognition and Catalysis by the Cofactor-Independent Dioxygenase DpgC.
Biochemistry, 46, 2007
|
|
7VF6
 
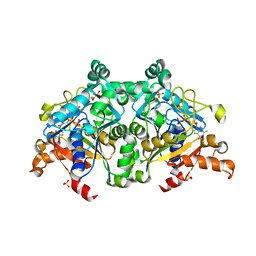 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
2L8L
 
 | |
4R89
 
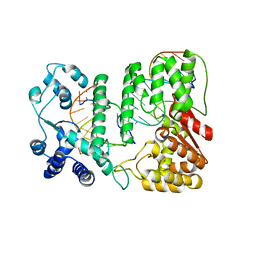 | | Crystal structure of paFAN1 - 5' flap DNA complex with Manganase | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
2OBS
 
 | |
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBR
 
 | | Crystal Structures of P Domain of Norovirus VA387 | | Descriptor: | Capsid protein | | Authors: | Cao, S, Lou, Z, Jiang, X, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2006-12-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of blood group trisaccharides by norovirus.
J.Virol., 81, 2007
|
|
2OBT
 
 | |
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
8JJB
 
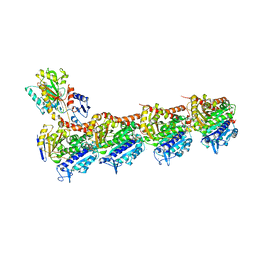 | | Crystal structure of T2R-TTL-Y61 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
5X6C
 
 | | Crystal structure of SepRS-SepCysE from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, O-phosphoserine--tRNA(Cys) ligase, SULFATE ION, ... | | Authors: | Chen, M, Kato, K, Yao, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural basis for tRNA-dependent cysteine biosynthesis
Nat Commun, 8, 2017
|
|
5X6B
 
 | |
8GZZ
 
 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | Descriptor: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
