6G43
 
 | | Crystal structure of SeMet-labeled mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4AN9
 
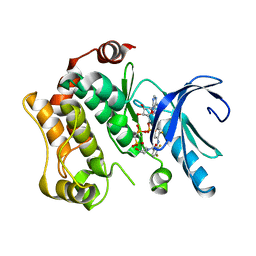 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
4A1F
 
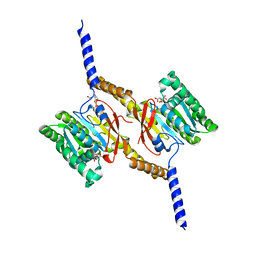 | | Crystal structure of C-terminal domain of Helicobacter pylori DnaB Helicase | | Descriptor: | CITRATE ANION, REPLICATIVE DNA HELICASE | | Authors: | Stelter, M, Kapp, U, Timmins, J, Terradot, L. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of a Dodecameric Bacterial Replicative Helicase.
Structure, 20, 2012
|
|
5LT1
 
 | |
1DJ3
 
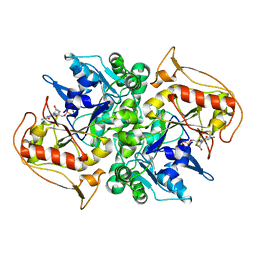 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
4AK5
 
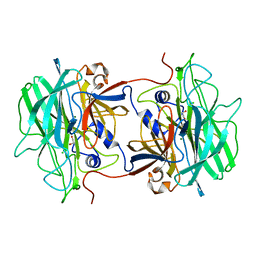 | | Native crystal structure of BpGH117 | | Descriptor: | 1,2-ETHANEDIOL, ANHYDRO-ALPHA-L-GALACTOSIDASE, CHLORIDE ION, ... | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
6GAA
 
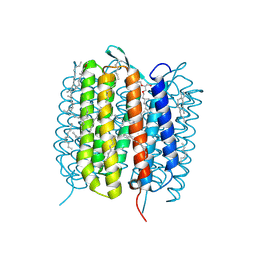 | | BACTERIORHODOPSIN, 430 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
1E1W
 
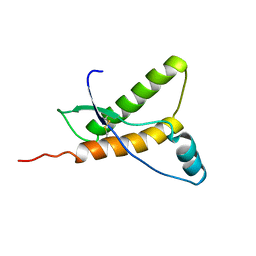 | | Human prion protein variant R220K | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-11 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6G6N
 
 | | Crystal structure of the computationally designed Tako8 protein in C2 | | Descriptor: | Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
4AJD
 
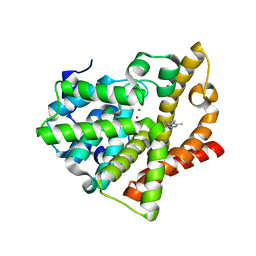 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | 2-ETHYL-4-METHYL-PHTHALAZIN-1-ONE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
3UEQ
 
 | | Crystal structure of amylosucrase from Neisseria polysaccharea in complex with turanose | | Descriptor: | 3-O-alpha-D-glucopyranosyl-D-fructose, Amylosucrase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guerin, F, Pizzut-Serin, S, Potocki-Veronese, G, Guillet, V, Mourey, L, Remaud-Simeon, M, Andre, I, Tranier, S. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Investigation of the Thermostability and Product Specificity of Amylosucrase from the Bacterium Deinococcus geothermalis.
J.Biol.Chem., 287, 2012
|
|
1DM1
 
 | | 2.0 A CRYSTAL STRUCTURE OF THE DOUBLE MUTANT H(E7)V, T(E10)R OF MYOGLOBIN FROM APLYSIA LIMACINA | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Federici, L, Savino, C, Musto, R, Travaglini-Allocatelli, C, Cutruzzola, F, Brunori, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering His(E7) affects the control of heme reactivity in Aplysia limacina myoglobin.
Biochem.Biophys.Res.Commun., 269, 2000
|
|
4AK6
 
 | | BpGH117_H302E mutant glycoside hydrolase | | Descriptor: | ANHYDRO-ALPHA-L-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
6GAE
 
 | | BACTERIORHODOPSIN, 560 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
8CX3
 
 | | Crystal structure of full-length mesothelin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhan, J, Esser, L, Xia, D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.609 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
1DOJ
 
 | | Crystal structure of human alpha-thrombin*RWJ-51438 complex at 1.7 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUGEN, ... | | Authors: | Recacha, R, Costanzo, M.J, Maryanoff, B.E, Carson, M, DeLucas, L, Chattopadhyay, D. | | Deposit date: | 1999-12-21 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human alpha-thrombin complexed with RWJ-51438 at 1.7 A: unusual perturbation of the 60A-60I insertion loop.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E2L
 
 | | Kinetics and crystal structure of the wild-type and the engineered Y101F mutant of Herpes simplex virus type 1 thymidine kinase interacting with (North)-methanocarba-thymidine | | Descriptor: | 1-[4-HYDROXY-5-(HYDROXYMETHYL)BICYCLO[3.1.0]HEX-2-YL]-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetics and Crystal Structure of the Wild-Type and the Engineered Y101F Mutant of Herpes Simplex Virus Type 1 Thymidine Kinase Interacting with (North)-Methanocarba-Thymidine
Biochemistry, 39, 2000
|
|
1DG3
 
 | | STRUCTURE OF HUMAN GUANYLATE BINDING PROTEIN-1 IN NUCLEOTIDE FREE FORM | | Descriptor: | PROTEIN (INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1) | | Authors: | Prakash, B, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 1999-11-23 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature, 403, 2000
|
|
6GBU
 
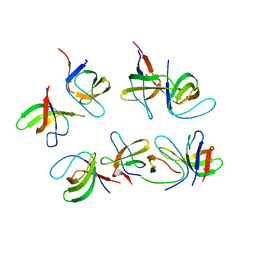 | | Crystal structure of the second SH3 domain of FCHSD2 (SH3-2) in complex with the fourth SH3 domain of ITSN1 (SH3d) | | Descriptor: | F-BAR and double SH3 domains protein 2, Intersectin-1 | | Authors: | Almeida-Souza, L, Frank, R, Garcia-Nafria, J, Colussi, A, Gunawardana, N, Johnson, C.M, Yu, M, Howard, G, Andrews, B, Vallis, Y, McMahon, H.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits.
Cell, 174, 2018
|
|
5MUR
 
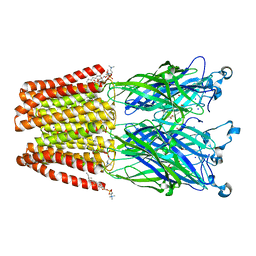 | | X-ray structure of the F14'A mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Fourati, Z, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
8W8O
 
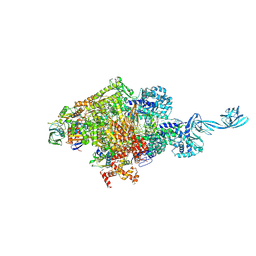 | |
5ZID
 
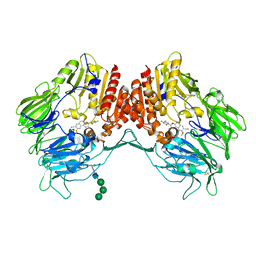 | | Crystal Structure of human DPP-IV in complex with HL2 | | Descriptor: | (2S,3R)-2-amino-9-methoxy-3-(2,4,5-trifluorophenyl)-2,3-dihydro-1H-naphtho[2,1-b]pyran-8-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Zhu, L, Li, H, Wu, F. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL2
To Be Published
|
|
6H0Q
 
 | | B1-type ACP domain from module 7 of MLSB | | Descriptor: | Type I modular polyketide synthase | | Authors: | Moretto, L, Heylen, R, Holroyd, N, Vance, S, Broadhurst, R.W. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Modular type I polyketide synthase acyl carrier protein domains share a common N-terminally extended fold.
Sci Rep, 9, 2019
|
|
5ZJS
 
 | | Structure of AbdB/Exd complex bound to a 'Blue14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
1DTY
 
 | | CRYSTAL STRUCTURE OF ADENOSYLMETHIONINE-8-AMINO-7-OXONANOATE AMINOTRANSFERASE WITH PYRIDOXAL PHOSPHATE COFACTOR. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Alexeev, D, Sawyer, L, Baxter, R.L, Alexeeva, M.V, Campopiano, D. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of adenosylmethionine-8-amino-7-oxonanoate aminotransferase with pyridoxal phosphate cofactor
to be published
|
|
