4YSJ
 
 | |
6QVD
 
 | |
6QV6
 
 | |
6QVB
 
 | |
5DDW
 
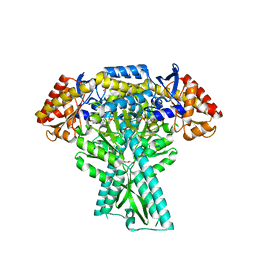 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
6QVU
 
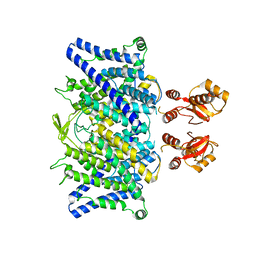 | |
6QVC
 
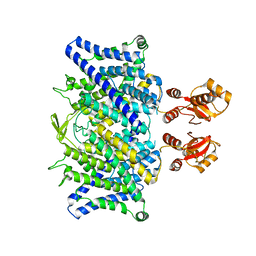 | |
7YRD
 
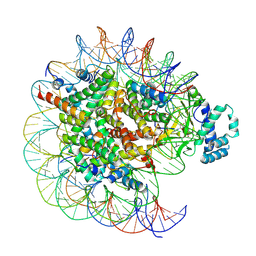 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7YRG
 
 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
5IQK
 
 | | Rm3 metallo-beta-lactamase | | Descriptor: | ZINC ION, beta-lactamase Rm3 | | Authors: | Salimraj, R, Spencer, J. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Rm3, a Subclass B3 Metallo-beta-Lactamase Identified from a Functional Metagenomic Study.
Antimicrob.Agents Chemother., 60, 2016
|
|
8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | Descriptor: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
6E5U
 
 | |
5DDS
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
3D34
 
 | | Structure of the F-spondin domain of mindin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Spondin-2 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-02-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
Embo J., 28, 2009
|
|
5JYK
 
 | | Deg9 crystal under 289K | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Zhang, L.X. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
5DDU
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
8IPS
 
 | |
8IPR
 
 | |
