4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
5UE7
 
 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
4DQ6
 
 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Putative pyridoxal phosphate-dependent transferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
5TCH
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - ligand-free form, TrpA-G66V mutant | | Descriptor: | FORMIC ACID, MALONATE ION, Tryptophan synthase alpha chain, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
4NF2
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis in complex with carbamoyl phosphate and L-norvaline | | Descriptor: | CHLORIDE ION, NORVALINE, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Handing, K, Cymborowski, M.T, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
5TCI
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, FORMIC ACID, MALONATE ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5T79
 
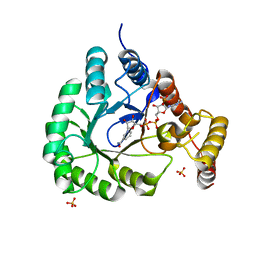 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | Descriptor: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
4EIV
 
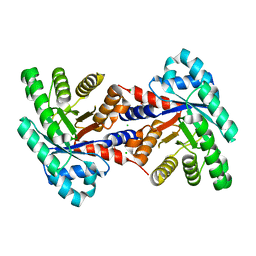 | | 1.37 Angstrom resolution crystal structure of apo-form of a putative deoxyribose-phosphate aldolase from Toxoplasma gondii ME49 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Deoxyribose-phosphate aldolase | | Authors: | Halavaty, A.S, Ruan, J, Minasov, G, Shuvalova, L, Ueno, A, Igarashi, M, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma gondii.
J.Mol.Biol., 427, 2015
|
|
4MA5
 
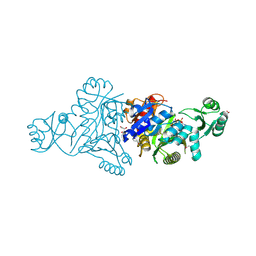 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
4ESE
 
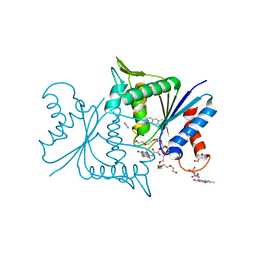 | | The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN. | | Descriptor: | DODECAETHYLENE GLYCOL, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of acyl carrier protein phosphodiesterase from Yersinia pestis CO92 in complex with FMN.
To be Published
|
|
4M9D
 
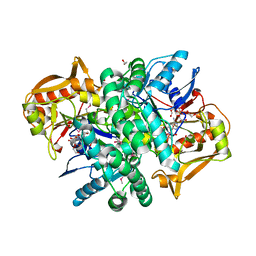 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
5UXA
 
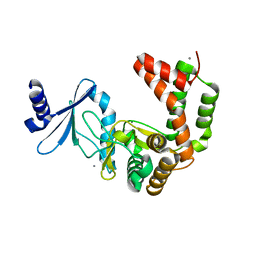 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
3MZZ
 
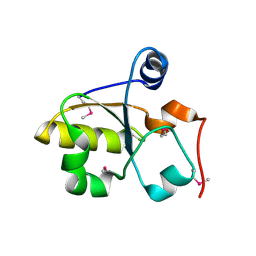 | | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus | | Descriptor: | Rhodanese-like domain protein | | Authors: | Kim, Y, Chruszcz, M, Minor, W, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus
To be Published
|
|
5UTT
 
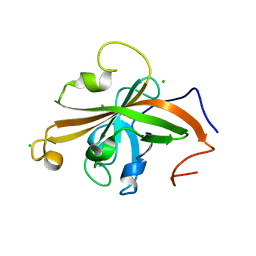 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4MYA
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110 | | Descriptor: | 4-{(1R)-1-[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]ethoxy}quinolin-2(1H)-one, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110
To be Published
|
|
5UXB
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5V0S
 
 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | Descriptor: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
4DE4
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-Id/APH(2")-IVa in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APH(2")-Id | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
5UCC
 
 | | Crystal structure of the ENTH domain of ENT2 from Candida albicans | | Descriptor: | CHLORIDE ION, CITRIC ACID, Potential epsin-like clathrin-binding protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the ENTH domain of ENT2 from Candida albicans
To Be Published
|
|
3P4E
 
 | | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae.
To be Published
|
|
5TCJ
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate and BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
4OIG
 
 | |
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4MFG
 
 | | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.
TO BE PUBLISHED
|
|
5VQB
 
 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
