3TEG
 
 | | Bacterial and Eukaryotic Phenylalanyl-tRNA Synthetases Catalyze Misaminoacylation of tRNAPhe with 3,4-Dihydroxy-L-Phenylalanine (L-Dopa) | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, Phenylalanyl-tRNA synthetase, mitochondrial | | Authors: | Moor, N, Klipcan, L, Safro, M. | | Deposit date: | 2011-08-14 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2044 Å) | | Cite: | Bacterial and Eukaryotic Phenylalanyl-tRNA Synthetases Catalyze Misaminoacylation of tRNA(Phe) with 3,4-Dihydroxy-L-Phenylalanine.
Chem.Biol., 18, 2011
|
|
1P3R
 
 | | Crystal structure of the phosphotyrosin binding domain(PTB) of mouse Disabled 1(Dab1) | | Descriptor: | Disabled homolog 2 | | Authors: | Yun, M, Keshvara, L, Park, C.G, Zhang, Y.M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.W. | | Deposit date: | 2003-04-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2.
J.Biol.Chem., 278, 2003
|
|
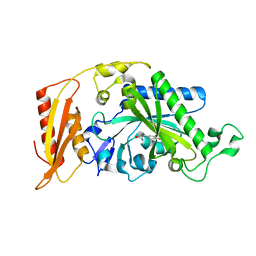
3T8K
 
 | |
1OOZ
 
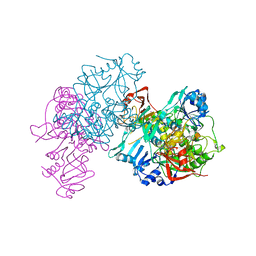 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
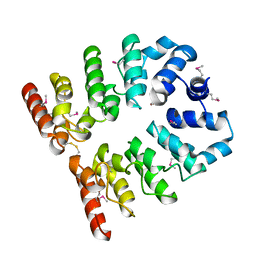
6GPD
 
 | | Crystal structure of the ligand-free form of domain 1 from TmArgBP | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
8C7C
 
 | | Double mutant V(M84)C/A(L278)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-14 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8RDQ
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8b | | Descriptor: | (5S)-3-(2,4-dichlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
3TFX
 
 | | Crystal structure of Orotidine 5'-phosphate decarboxylase from Lactobacillus acidophilus | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-16 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Orotidine 5'-phosphate decarboxylase from Lactobacillus acidophilus
To be Published
|
|
8VR9
 
 | | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Maso, L, Bang, I, Koide, S. | | Deposit date: | 2024-01-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8C6K
 
 | | Double mutant A(L53)C/I(L64)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C87
 
 | | Double mutant A(L172)C/L(L246)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Yukhimchuk, D. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C5X
 
 | | Double mutant A(L37)C/S(L99)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
6GI2
 
 | |
8VRA
 
 | | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | | Descriptor: | AMG 510 (bound form), Beta-2-microglobulin, GTPase KRas, ... | | Authors: | Maso, L, Bang, I, Koide, S. | | Deposit date: | 2024-01-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8C88
 
 | | Double mutant G(M19)C/T(L214)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Yukhimchuk, D. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
1OUX
 
 | | LecB (PA-LII) sugar-free | | Descriptor: | CALCIUM ION, SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
5MOB
 
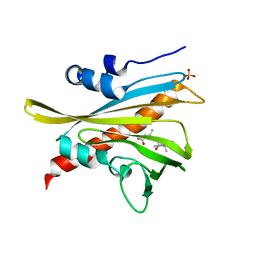 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, SULFATE ION, SlPYL1_ABA | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
6GI6
 
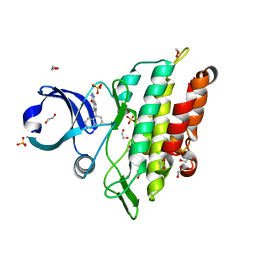 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6GIH
 
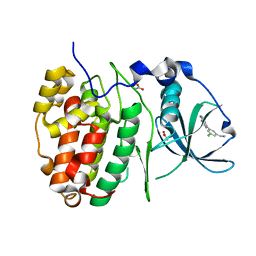 | | Crystal Structure of CK2alpha with CAM187 bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, [3-chloranyl-5-(1~{H}-indol-4-yl)phenyl]methanamine | | Authors: | Brear, P, Iegre, J, North, A, De Fusco, C, Georgiou, K, Lubin, A, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel non-ATP competitive small molecules targeting the CK2 alpha / beta interface.
Bioorg. Med. Chem., 26, 2018
|
|
1FFM
 
 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII (FUCOSYLATED AT SER-60), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (Blood Coagulation Factor VII), alpha-L-fucopyranose | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
4CR5
 
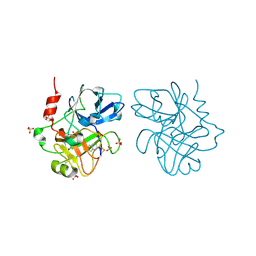 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-chloroquinolin-2(1H)-one, COAGULATION FACTOR XIA, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-25 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CRC
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | (2S)-2-[[(E)-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-3-phenyl-N-[4-(1H-1,2,3,4-tetrazol-5-yl)phenyl]propanamide, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
8VI8
 
 | | Engineered glutamine binding protein and a cobaloxime ligand - no GLN bound | | Descriptor: | AZIDOBIS (DIMETHYLGLYOXIMATO) PYRIDINECOBALT, Amino acid ABC transporter substrate-binding protein, SODIUM ION, ... | | Authors: | Boggs, D.G, Fatima, S, Olshansky, L, Bridwell-Rabb, J. | | Deposit date: | 2024-01-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Conformation-Dependent Hydrogen-Bonding Interactions in a Switchable Artificial Metalloprotein.
Biochemistry, 63, 2024
|
|
5M6I
 
 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
