3IEU
 
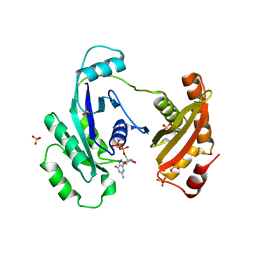 | | Crystal Structure of ERA in Complex with GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GTP-binding protein era, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IEV
 
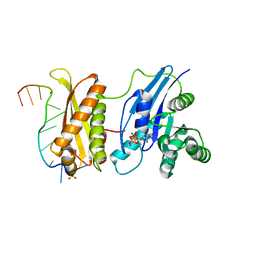 | | Crystal Structure of ERA in Complex with MgGNP and the 3' End of 16S rRNA | | Descriptor: | 5'-R(P*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3', GTP-binding protein era, MAGNESIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IO2
 
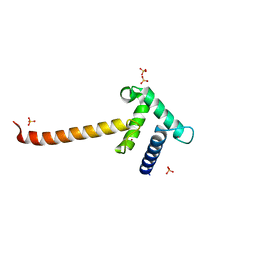 | | Crystal structure of the Taz2 domain of p300 | | Descriptor: | Histone acetyltransferase p300, SULFATE ION, ZINC ION | | Authors: | Miller, M, Dauter, Z, Wlodawer, A. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Taz2 domain of p300: insights into ligand binding.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3T92
 
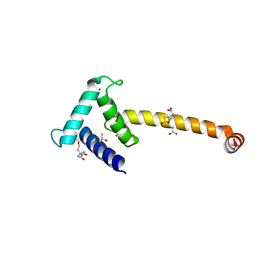 | | Crystal structure of the Taz2:C/EBPepsilon-TAD chimera protein | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ACETONE, HISTONE ACETYLTRANSFERASE P300 TAZ2-CCAAT/ENHANCER-BINDING PROTEIN EPSILON, ... | | Authors: | Bhaumik, P, Maria, M. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into interactions of C/EBP transcriptional activators with the Taz2 domain of p300.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
2K5S
 
 | | YmoA | | Descriptor: | Modulating protein ymoA | | Authors: | McFeeters, R.L, Byrd, R. | | Deposit date: | 2008-06-30 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The high-precision solution structure of Yersinia modulating
protein YmoA provides insight into interaction with H-NS
Biochemistry, 46, 2007
|
|
2K8F
 
 | | Structural Basis for the Regulation of p53 Function by p300 | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
6PC1
 
 | | Crystal structure of Helicobacter pylori PPX/GppA (E143A) in complex with ppGpp | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC0
 
 | | Crystal structure of Helicobacter pylori PPX/GppA | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, D-MALATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PBZ
 
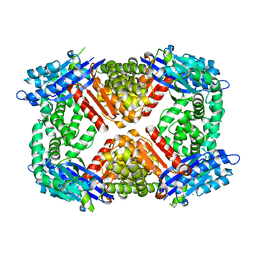 | | Crystal structure of Escherichia coli GppA | | Descriptor: | CHLORIDE ION, Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase | | Authors: | Song, H, Shaw, G.X, Wang, C, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
3EE1
 
 | |
6PC3
 
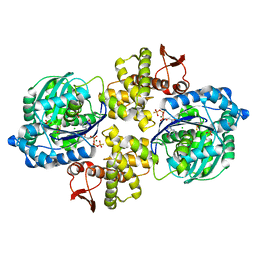 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GSP | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC2
 
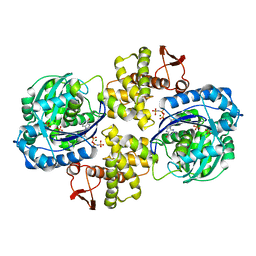 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GNP | | Descriptor: | Guanosine pentaphosphate phosphohydrolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
3FTD
 
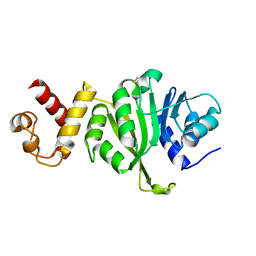 | |
3FTF
 
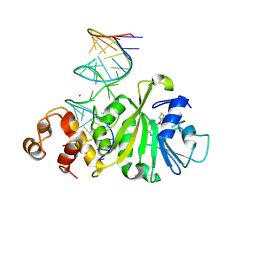 | | Crystal structure of A. aeolicus KsgA in complex with RNA and SAH | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase, POTASSIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
3FTC
 
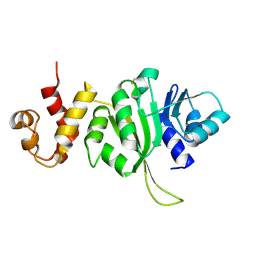 | |
3FTE
 
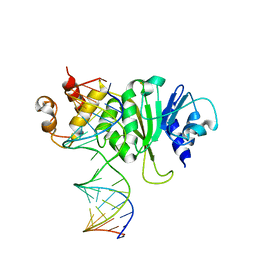 | | Crystal structure of A. aeolicus KsgA in complex with RNA | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
8DVH
 
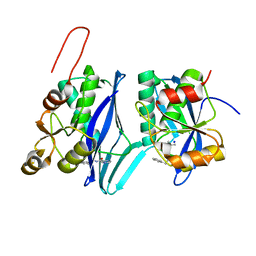 | | Crystal structure of ATP-dependent Lon protease from Bacillus subtillis (BsLonBA) | | Descriptor: | Lon protease 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, SODIUM ION | | Authors: | Sekula, B, Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique Structural Fold of LonBA Protease from Bacillus subtilis, a Member of a Newly Identified Subfamily of Lon Proteases.
Int J Mol Sci, 23, 2022
|
|
1I4S
 
 | |
1JFZ
 
 | |
7R97
 
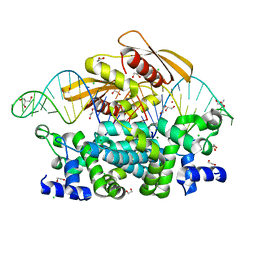 | | Crystal structure of postcleavge complex of Escherichia coli RNase III | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Dharavath, S, Shaw, G.X, Ji, X. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg 2+ -ion catalysis.
Rna Biol., 19, 2022
|
|
6W4R
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ633p | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyrazin-3-yl]amino}benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
6W7J
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ635p | | Descriptor: | 1,2-ETHANEDIOL, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
7KDO
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-73 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
7KDR
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]sulfonyl}-5'-deoxyadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.488 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
1Z0V
 
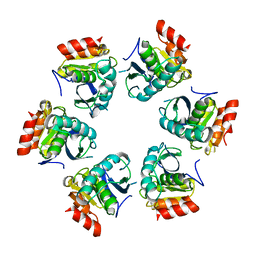 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Dauter, Z, Botos, I, LaRonde-LeBlanc, N, Wlodawer, A. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pathological crystallography: case studies of several unusual macromolecular crystals.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
