6UAG
 
 | |
6U2Z
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, 1,2-ETHANEDIOL, COPPER (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions
To Be Published
|
|
6UAC
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with cadmium and hydrolyzed moxolactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, CADMIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with cadmium and hydrolyzed moxolactam
To Be Published
|
|
6UAP
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound
To be Published
|
|
6UG4
 
 | |
6U0Z
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed penicillin G | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed penicillin G.
To Be Published
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
8U00
 
 | | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides
To Be Published
|
|
8U12
 
 | |
8TTP
 
 | | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-14 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam
to be published
|
|
8UW6
 
 | | Acetylornithine deacetylase from Escherichia coli, di-zinc form. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetylornithine deacetylase, ... | | Authors: | Osipiuk, J, Endres, M, Kelley, E, Becker, D.P, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetylornithine deacetylase from Escherichia coli, di-zinc form.
To Be Published
|
|
2FSQ
 
 | | Crystal Structure of the Conserved Protein of Unknown Function ATU0111 from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Atu0111 protein | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Atu0111 from Agrobacterium tumefaciens str. C58
To be Published
|
|
3LG2
 
 | | A Ykr043C/ fructose-1,6-bisphosphate product complex following ligand soaking | | Descriptor: | PHOSPHATE ION, Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
2FSR
 
 | | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, acetyltransferase | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58
To be Published
|
|
2G39
 
 | | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acetyl-CoA hydrolase | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa
To be Published
|
|
2FRE
 
 | | The crystal structure of the oxidoreductase containing FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H-flavin oxidoreductase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-19 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the oxidoreductase containing FMN from Agrobacterium tumefaciens
To be Published
|
|
2GE3
 
 | | Crystal structure of Probable acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | ACETYL COENZYME *A, probable acetyltransferase | | Authors: | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Probable acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2FXA
 
 | | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis. | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis.
TO BE PUBLISHED
|
|
2GEN
 
 | | Structural Genomics, the crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | probable transcriptional regulator | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-20 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
2GFN
 
 | | Crystal structure of HTH-type transcriptional regulator pksA related protein from Rhodococcus sp. RHA1 | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator pksA related protein | | Authors: | Nocek, B, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of HTH-type transcriptional regulator pksA related protein from Rhodococcus sp. RHA1
To be Published
|
|
2G84
 
 | | Cytidine and deoxycytidylate deaminase zinc-binding region from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of Cytidine and deoxycytidylate deaminase zinc-binding region from Nitrosomonas europaea.
To be Published
|
|
2G8Y
 
 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
2G7L
 
 | | Crystal structure of putative transcription regulator SCO7704 from Streptomyces coelicor | | Descriptor: | TetR-family transcriptional regulator | | Authors: | Ezersky, A, Lunin, V.V, Skarina, T, Wierzbicka, M, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative transcription regulator SCO7704 from Streptomyces coelicor
To be Published
|
|
2G9I
 
 | | Crystal structure of homolog of F420-0:gamma-Glutamyl Ligase from Archaeoglobus fulgidus Reveals a Novel Fold. | | Descriptor: | F420-0:gamma-glutamyl ligase | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-04-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an amide bond forming F(420):gamma-glutamyl ligase from Archaeoglobus fulgidus -- a member of a new family of non-ribosomal peptide synthases.
J.Mol.Biol., 372, 2007
|
|
3CIT
 
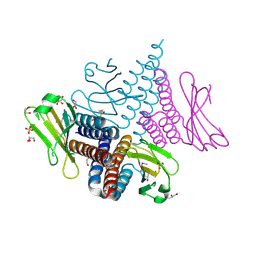 | | Crystal structure of the GAF domain of a putative sensor histidine kinase from Pseudomonas syringae pv. tomato | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GAF domain of a putative sensor histidine kinase from Pseudomonas syringae pv. tomato
To be Published
|
|
