5VSK
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VSB
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-{[4-hydroxy-1-(3-phenylpropanoyl)piperidin-4-yl]methyl}quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
3ZNR
 
 | | HDAC7 bound with inhibitor TMP269 | | Descriptor: | HISTONE DEACETYLASE 7, N-{[4-(4-phenyl-1,3-thiazol-2-yl)tetrahydro-2H-pyran-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | Authors: | Lobera, m, madauss, k, pohlhaus, d, trump, r, nolan, m. | | Deposit date: | 2013-02-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
5BRW
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BRU
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
1XJD
 
 | |
4COX
 
 | |
3PGH
 
 | |
1HCQ
 
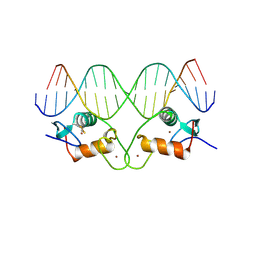 | | THE CRYSTAL STRUCTURE OF THE ESTROGEN RECEPTOR DNA-BINDING DOMAIN BOUND TO DNA: HOW RECEPTORS DISCRIMINATE BETWEEN THEIR RESPONSE ELEMENTS | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP*CP*CP*T P*G)-3'), DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP*CP*CP*T P*G)-3'), PROTEIN (ESTROGEN RECEPTOR), ... | | Authors: | Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1995-01-04 | | Release date: | 1995-11-23 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements.
Cell(Cambridge,Mass.), 75, 1993
|
|
2V37
 
 | | Solution structure of the N-terminal extracellular domain of human T- cadherin | | Descriptor: | CADHERIN-13 | | Authors: | Dames, S.A, Bang, E.J, Ahrens, T, Haeussinger, D, Grzesiek, S. | | Deposit date: | 2007-06-13 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the low adhesive capacity of human T-cadherin from the NMR structure of Its N-terminal extracellular domain.
J. Biol. Chem., 283, 2008
|
|
6S07
 
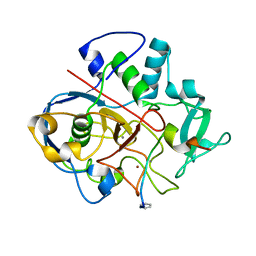 | | Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen. | | Descriptor: | Abz-ALA-THR-THR-PRO-LEU-CYS-GLY-PRO-SER-ARG-ALA-SER-ILE-LEU-SER-GLY-ARG, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Leisinger, F, Miarzlou, D.A, Seebeck, F.P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Chem Sci, 10, 2019
|
|
8BU2
 
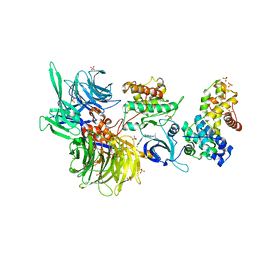 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUI
 
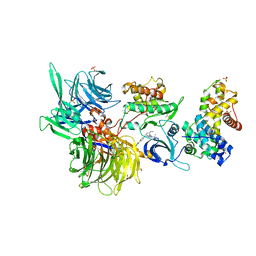 | | Structure of DDB1 bound to DRF-053-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)amino]purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUA
 
 | | Structure of DDB1 bound to 919278-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-~{N}-(1~{H}-benzimidazol-2-yl)-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanamide, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.193 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUR
 
 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUF
 
 | | Structure of DDB1 bound to Z12-engaged CDK12-cyclin K | | Descriptor: | 2-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylmethyl)-6,7-dimethoxy-3~{H}-quinazolin-4-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU4
 
 | | Structure of DDB1 bound to DS22-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU1
 
 | | Structure of DDB1 bound to DS17-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUJ
 
 | | Structure of DDB1 bound to DS06-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-(octylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUO
 
 | | Structure of DDB1 bound to DS24-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[(3-fluoranyl-4-pyridin-2-yl-phenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU9
 
 | | Structure of DDB1 bound to roscovitine-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU6
 
 | | Structure of DDB1 bound to DS55-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUP
 
 | | Structure of DDB1 bound to DS30-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[3-(3-methylphenyl)propylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
