2CW4
 
 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-16 | | Release date: | 2005-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
2DB7
 
 | | Crystal structure of hypothetical protein MS0332 | | Descriptor: | Hairy/enhancer-of-split related with YRPW motif 1 | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical protein MS0332
To be Published
|
|
6HBA
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
2DTC
 
 | | Crystal structure of MS0666 | | Descriptor: | RAL GUANINE NUCLEOTIDE EXCHANGE FACTOR RALGPS1A | | Authors: | Wang, H, Kishishita, S, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-12 | | Release date: | 2007-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MS0666
To be Published
|
|
2CVL
 
 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
6HBC
 
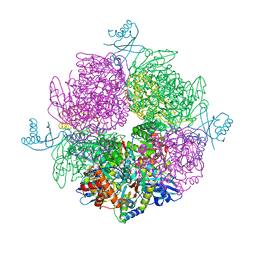 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
2F5H
 
 | | Solution structure of the alpha-domain of human Metallothionein-3 | | Descriptor: | CADMIUM ION, Metallothionein-3 | | Authors: | Wang, H, Zhang, Q, Cai, B, Li, H.Y, Sze, K.H, Huang, Z.X, Wu, H.M, Sun, H.Z. | | Deposit date: | 2005-11-25 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of human metallothionein-3 (MT-3)
Febs Lett., 580, 2006
|
|
6HBB
 
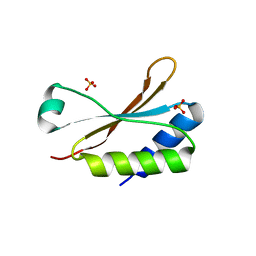 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
2R8Q
 
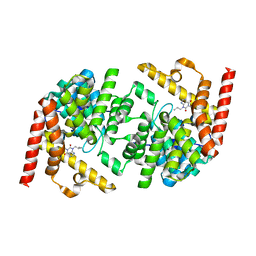 | | Structure of LmjPDEB1 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, Class I phosphodiesterase PDEB1, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Geng, J, Kunz, S, Seebeck, T, Ke, H. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors.
Mol.Microbiol., 66, 2007
|
|
3FG7
 
 | |
6CM3
 
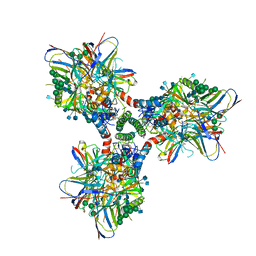 | | BG505 SOSIP in complex with sCD4, 17b, 8ANC195 | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2018-03-02 | | Release date: | 2018-10-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion.
Cell Host Microbe, 24, 2018
|
|
6BYF
 
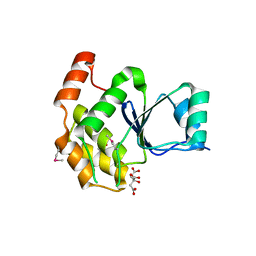 | |
3HBT
 
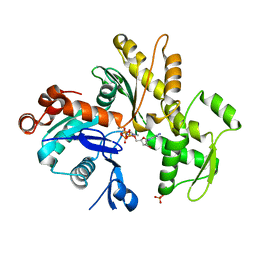 | | The structure of native G-actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Wang, H, Robinson, R.C, Burtnick, L.D. | | Deposit date: | 2009-05-05 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of native G-actin
Cytoskeleton (Hoboken), 67, 2010
|
|
2RID
 
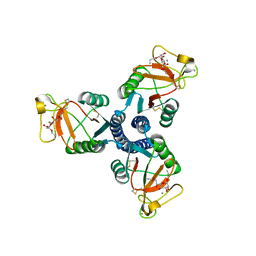 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with Allyl 7-O-carbamoyl-L-glycero-D-manno-heptopyranoside | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, prop-2-en-1-yl 7-O-carbamoyl-L-glycero-alpha-D-manno-heptopyranoside | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
7CGC
 
 | |
7CGD
 
 | | Silver-bound E.coli malate dehydrogenase | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm.
Chem Sci, 11, 2020
|
|
7CB0
 
 | |
7CB5
 
 | |
7CB6
 
 | |
7MOJ
 
 | |
7MOM
 
 | |
7MOF
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 6-diphosphoinositol 1,2,3,4,5-pentakisphosphate 6-InsP7 | | Descriptor: | (1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, INOSITOL HEXAKISPHOSPHATE, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOH
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5PP-InsP4) and phosphate in conformation B (Pi(B)) | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, PHOSPHATE ION, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOE
 
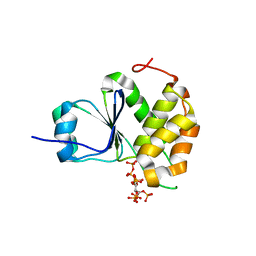 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1)Cys150Ser in complex with 5-diphosphoinositol 1,2,3,4,6-pentakisphosphate (5-InsP7) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOL
 
 | |
