2GVX
 
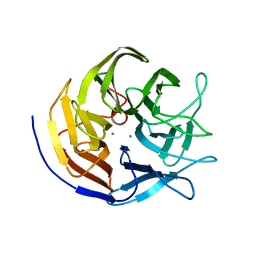 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
4PED
 
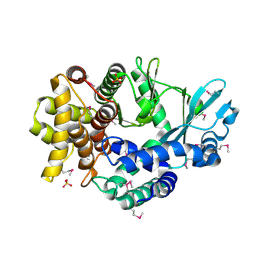 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
4RZA
 
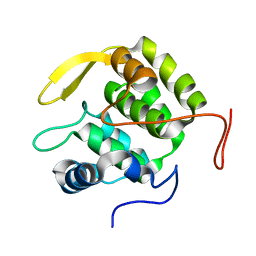 | | Pre-mRNA-splicing factor 38A AS 1-205 | | Descriptor: | Pre-mRNA-splicing factor 38A | | Authors: | Schuetze, T, Ulrich, A, Wahl, M.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The N-terminal domain of the unusual SR protein hPrp38 is an interaction hub in the spliceosome
To be Published
|
|
8PKM
 
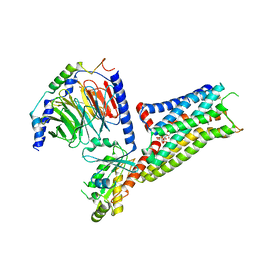 | | Befiradol-bound serotonin 5-HT1A receptor - Gi Protein Complex | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, (3-chloranyl-4-fluoranyl-phenyl)-[4-fluoranyl-4-[[(5-methylpyridin-2-yl)methylamino]methyl]piperidin-1-yl]methanone, 5-hydroxytryptamine receptor 1A, ... | | Authors: | Schneider, J, Gmeiner, P, Hove, T.T, Rasmussen, T, Boettcher, B. | | Deposit date: | 2023-06-27 | | Release date: | 2024-05-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a functionally selective serotonin receptor (5-HT 1A R) agonist for the treatment of pain.
Sci Adv, 11, 2025
|
|
8PJK
 
 | | ST171-bound serotonin 5-HT1A receptor - Gi Protein Complex | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 5-hydroxytryptamine receptor 1A, 6-[3-[2-(2-methoxyphenoxy)ethylamino]propoxy]-4~{H}-1,4-benzoxazin-3-one, ... | | Authors: | Schneider, J, Gmeiner, P, Rasmussen, T, Boettcher, B. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Discovery of a functionally selective serotonin receptor (5-HT 1A R) agonist for the treatment of pain.
Sci Adv, 11, 2025
|
|
6HFV
 
 | | Mycobacterium tuberculosis DprE1 in complex with CMP2 | | Descriptor: | 8-(oxidanylamino)-2-piperidin-1-yl-6-(trifluoromethyl)-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chung, C. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HF3
 
 | | M tuberculosis DprE1 in complex with a covalently bound nitrobenzothiazinone | | Descriptor: | 2-(4-(cyclohexylmethyl)piperazin-1-yl)-8-nitro-6-(trifluoromethyl)-4H-benzo[e][1,3]thiazin-4-one, bound form, Decaprenylphosphoryl-beta-D-ribose oxidase, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HF0
 
 | | M. tuberculosis DprE1 covalently bound to a nitrobenzoxacinone. | | Descriptor: | 2-[(2~{S},6~{R})-2,6-dimethylpiperidin-1-yl]-8-nitro-6-(trifluoromethyl)-1,3-benzoxazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HEZ
 
 | | M tuberculosis DprE1 in complex with BTZ043 | | Descriptor: | 8-(hydroxyamino)-2-[(2S)-2-methyl-1,4-dioxa-8-azaspiro[4.5]dec-8-yl]-6-(trifluoromethyl)-4H-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HFW
 
 | | Mycobacterium tuberculosis DprE1 in complex with CMP1 | | Descriptor: | 8-(oxidanylamino)-2-piperidin-1-yl-6-(trifluoromethyl)-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chung, C. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
9GL2
 
 | | Befiradol-bound serotonin 5-HT1A receptor - Gs Protein Complex | | Descriptor: | (2R)-1-(heptadecanoyloxy)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, (3-chloranyl-4-fluoranyl-phenyl)-[4-fluoranyl-4-[[(5-methylpyridin-2-yl)methylamino]methyl]piperidin-1-yl]methanone, 5-hydroxytryptamine receptor 1A, ... | | Authors: | Schneider, J, Gmeiner, P, Boettcher, B, Rasmussen, T. | | Deposit date: | 2024-08-26 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of a functionally selective serotonin receptor (5-HT 1A R) agonist for the treatment of pain.
Sci Adv, 11, 2025
|
|
3UVR
 
 | | Human p38 MAP Kinase in Complex with KM064 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{3-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
1KPY
 
 | |
1KPZ
 
 | |
6TNA
 
 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
8F2Y
 
 | |
3F21
 
 | | Crystal structure of Zalpha in complex with d(CACGTG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DAP*DCP*DGP*DTP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3EYI
 
 | |
3F23
 
 | | Crystal structure of Zalpha in complex with d(CGGCCG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DCP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3F22
 
 | | Crystal structure of Zalpha in complex with d(CGTACG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DTP*DAP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
5EBI
 
 | | Crystal structure of a DNA-RNA chimera in complex with Ba2+ ions: a case of unusual multi-domain twinning | | Descriptor: | BARIUM ION, DNA/RNA (5'-D(*C)-R(P*G)-D(P*C)-R(P*G)-D(P*C)-R(P*G)-3') | | Authors: | Gilski, M, Drozdzal, P, Kierzek, R, Jaskolski, M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3L2M
 
 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
3L2L
 
 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Limit Dextrin and Oligosaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
3IRQ
 
 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1D53
 
 | | CRYSTAL STRUCTURE AT 1.5 ANGSTROMS RESOLUTION OF D(CGCICICG), AN OCTANUCLEOTIDE CONTAINING INOSINE, AND ITS COMPARISON WITH D(CGCG) AND D(CGCGCG) STRUCTURES | | Descriptor: | DNA (5'-D(*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*IP*CP*IP*CP*G)-3') | | Authors: | Kumar, V.D, Harrison, R.W, Andrews, L.C, Weber, I.T. | | Deposit date: | 1992-11-05 | | Release date: | 1993-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures.
Biochemistry, 31, 1992
|
|
