6K6K
 
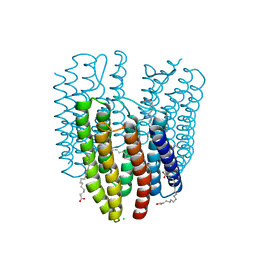 | | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens | | Descriptor: | CHLORIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer (N63A/P118A) Mastigocladopsis repens
To Be Published
|
|
6K6J
 
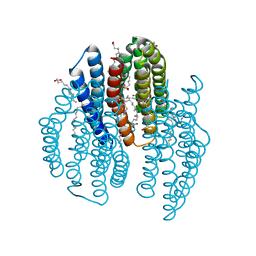 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | Descriptor: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | Descriptor: | RNA-binding protein 3 | | Authors: | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
7FCI
 
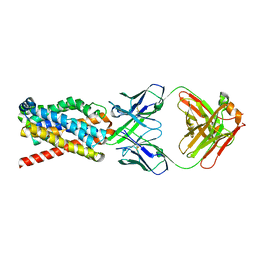 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
1G0V
 
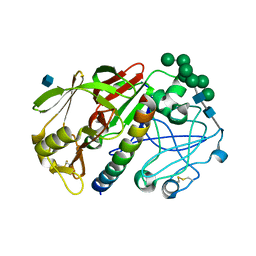 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
2AQU
 
 | | Structure of HIV-1 protease bound to atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, HIV-1 Protease | | Authors: | Clemente, J.C, Coman, R.M, Thiaville, M.M, Janka, L.K, Jeung, J.A, Nukoolkarn, S, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Leelamanit, W, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of HIV-1 CRF_01 A/E protease inhibitor resistance: structural determinants for maintaining sensitivity and developing resistance to atazanavir.
Biochemistry, 45, 2006
|
|
6B16
 
 | | P21-activated kinase 1 in complex with a 4-azaindole inhibitor | | Descriptor: | N~4~-(5-cyclopropyl-1H-pyrazol-3-yl)-N~2~-[(1S)-1-(1H-pyrrolo[3,2-b]pyridin-5-yl)ethyl]pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Rouge, L, Wang, W. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and evaluation of a series of 4-azaindole-containing p21-activated kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
2LGD
 
 | |
2MVF
 
 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4MLE
 
 | | Human Glucokinase in Complex with Novel Amino Thiazole Activator | | Descriptor: | 3-(benzyloxy)-N-(4-methyl-1,3-thiazol-2-yl)pyridin-2-amine, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W.C. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a New Class of Glucokinase Activators through Structure-Based Design.
J.Med.Chem., 56, 2013
|
|
4MLH
 
 | |
5BMS
 
 | |
1AYM
 
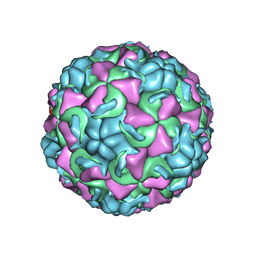 | | HUMAN RHINOVIRUS 16 COAT PROTEIN AT HIGH RESOLUTION | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle.
Structure, 5, 1997
|
|
6G4Z
 
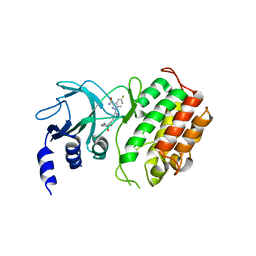 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 2f | | Descriptor: | 5-fluoranyl-1-[4-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrol-3-yl]ethynyl]pyridin-2-yl]indazole-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Leonardo-Silvestre, H, McEwan, P.A, Hymowitz, S.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
6G4Y
 
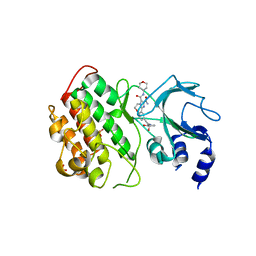 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 1a | | Descriptor: | 10-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrolidin-3-yl]ethynyl]-~{N}3-(oxan-4-yl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Hole, A.J, Hymowitz, S.G, McEwan, P.A. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
1JDQ
 
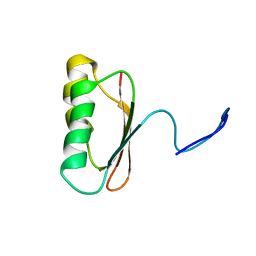 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JCU
 
 | |
1JRM
 
 | |
1JW2
 
 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1V3A
 
 | |
2WCV
 
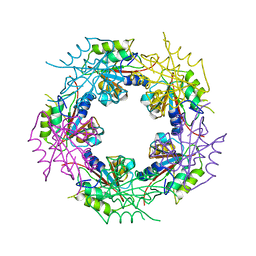 | | Crystal structure of bacterial FucU | | Descriptor: | L-FUCOSE MUTAROTASE, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
2WCU
 
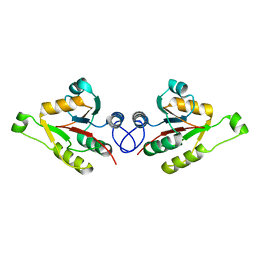 | | Crystal structure of mammalian FucU | | Descriptor: | PROTEIN FUCU HOMOLOG, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
4PPZ
 
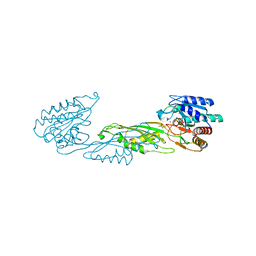 | | Crystal structure of zinc-bound succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 | | Descriptor: | PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
