1F7X
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN ZIPA | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Moy, F.J, Glasfeld, E, Mosyak, L, Powers, R. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ZipA, a crucial component of Escherichia coli cell division.
Biochemistry, 39, 2000
|
|
1FFV
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
1FI9
 
 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
2JRI
 
 | | Solution structure of the Josephin domain of Ataxin-3 in complex with ubiquitin molecule. | | Descriptor: | Ataxin-3, UBC protein | | Authors: | Nicastro, G, Masino, L, Esposito, V, Menon, R, Pastore, A. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Understanding the plasticity of the ubiquitin-protein recognition code: the josephin domain of ataxin-3 is a diubiquitin binding motif
To be Published
|
|
8TRO
 
 | | Rod from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
1F3Z
 
 | | IIAGLC-ZN COMPLEX | | Descriptor: | GLUCOSE-SPECIFIC PHOSPHOCARRIER, ZINC ION | | Authors: | Feese, M, Comolli, L, Meadow, N, Roseman, S, Remington, S.J. | | Deposit date: | 1997-10-09 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural studies of the Escherichia coli signal transducing protein IIAGlc: implications for target recognition.
Biochemistry, 36, 1997
|
|
1F9C
 
 | | CRYSTAL STRUCTURE OF MLE D178N VARIANT | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE I) | | Authors: | Kajander, T, Lehtio, L, Kahn, P.C, Goldman, A. | | Deposit date: | 2000-07-10 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Buried charged surface in proteins.
Structure Fold.Des., 8, 2000
|
|
1FCV
 
 | | CRYSTAL STRUCTURE OF BEE VENOM HYALURONIDASE IN COMPLEX WITH HYALURONIC ACID TETRAMER | | Descriptor: | HYALURONOGLUCOSAMINIDASE, alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid | | Authors: | Markovic-Housley, Z, Miglierini, G, Soldatova, L, Rizkallah, P.J, Mueller, U, Schirmer, T. | | Deposit date: | 2000-07-19 | | Release date: | 2001-10-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of hyaluronidase, a major allergen of bee venom.
Structure Fold.Des., 8, 2000
|
|
1FFU
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
8TNL
 
 | |
3CXF
 
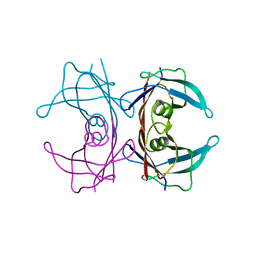 | | Crystal structure of transthyretin variant Y114H | | Descriptor: | Transthyretin | | Authors: | Cendron, L, Zanotti, G, Folli, C, Alfieri, B, Pasquato, N, Berni, R. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
8TOA
 
 | | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H7.HK2 Neutralizing Antibody Heavy Chain, H7.HK2 Neutralizing Antibody Light Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Wu, X, Shapiro, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK1
To Be Published
|
|
1FQ2
 
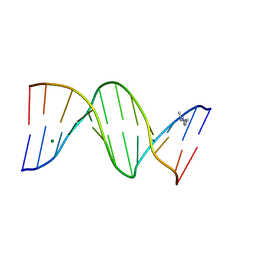 | | CRYSTAL STRUCTURE ANALYSIS OF THE POTASSIUM FORM OF B-DNA DODECAMER CGCGAATTCGCG | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Williams, L.D, Sines, C.C, McFail-Isom, L, Howerton, S.B, VanDerveer, D. | | Deposit date: | 2000-09-01 | | Release date: | 2000-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cations Mediate B-DNA Conformational Heterogeneity
J.Am.Chem.Soc., 122, 2000
|
|
3D74
 
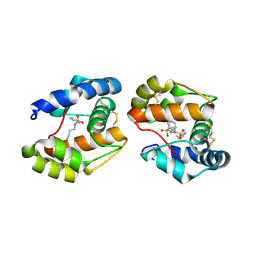 | | Crystal structure of a pheromone binding protein mutant D35A, from Apis mellifera, soaked at pH 5.5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
8T01
 
 | |
1FOF
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 | | Descriptor: | BETA LACTAMASE OXA-10, COBALT (II) ION, SULFATE ION | | Authors: | Paetzel, M, Danel, F, de Castro, L, Mosimann, S.C, Page, M.G.P, Strynadka, N.C.J. | | Deposit date: | 2000-08-28 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the class D beta-lactamase OXA-10.
Nat.Struct.Biol., 7, 2000
|
|
8T7C
 
 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLCg2 reveal dynamic inter-domain recognitions in autoinhibition
To Be Published
|
|
1FRQ
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
1D9K
 
 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
1CK3
 
 | | N276D MUTANT OF ESCHERICHIA COLI TEM-1 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE | | Authors: | Swaren, P, Maveyraud, L, Samama, J.P. | | Deposit date: | 1999-04-27 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray structure of the Asn276Asp variant of the Escherichia coli TEM-1 beta-lactamase: direct observation of electrostatic modulation in resistance to inactivation by clavulanic acid.
Biochemistry, 38, 1999
|
|
3D78
 
 | | Dimeric crystal structure of a pheromone binding protein mutant D35N, from apis mellifera, at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7XB5
 
 | |
1CVL
 
 | | CRYSTAL STRUCTURE OF BACTERIAL LIPASE FROM CHROMOBACTERIUM VISCOSUM ATCC 6918 | | Descriptor: | CALCIUM ION, TRIACYLGLYCEROL HYDROLASE | | Authors: | Lang, D.A, Hofmann, B, Haalck, L, Hecht, H.-J, Spener, F, Schmid, R.D, Schomburg, D. | | Deposit date: | 1997-01-09 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a bacterial lipase from Chromobacterium viscosum ATCC 6918 refined at 1.6 angstroms resolution.
J.Mol.Biol., 259, 1996
|
|
