6INK
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
8JB5
 
 | |
8X2I
 
 | |
8X2H
 
 | |
9M3U
 
 | | Crystal structure of human pyruvate dehydrogenase kinase isoform 2 in complex with ATP competitive inhibitor 24 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-N-[(3R)-1-[(3S)-3-ethyl-7-(ethylamino)-5-fluoranyl-2-oxidanylidene-1H-indol-3-yl]piperidin-3-yl]-2-azaspiro[3.3]heptane-6-carboxamide, GLYCEROL, ... | | Authors: | Xu, Z.H, Chen, S, Han, L. | | Deposit date: | 2025-03-03 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of ATP competitive PDHK1/2 dual inhibitors.
Bioorg.Med.Chem.Lett., 122, 2025
|
|
9M3R
 
 | |
9M3T
 
 | |
9M3O
 
 | |
9M3P
 
 | |
9LHJ
 
 | | UBE2N/UBE2V2 complexed with a covalent inhibitor | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Li, S, Wu, X, Zhou, L, Lu, X. | | Deposit date: | 2025-01-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9LG2
 
 | | Phosphoglycerate mutase 1 complexed with a covalent inhibitor | | Descriptor: | CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Li, S, Wu, X, Zhou, L, Lu, X. | | Deposit date: | 2025-01-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9LVG
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with Robinetin | | Descriptor: | 3,7-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of RIPK3 as a target of flavonoids for anti-necroptosis in vitro.
Bioorg.Chem., 161, 2025
|
|
9LVH
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with Tricetin | | Descriptor: | 5,7-DIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of RIPK3 as a target of flavonoids for anti-necroptosis in vitro.
Bioorg.Chem., 161, 2025
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9L3Z
 
 | | Cryo-EM structure of the inactive chemokine-like receptor 1 (CMKLR1) | | Descriptor: | Chemerin-like receptor 1,Soluble cytochrome b562, LRH7-C2, The heavy chain of anti-BRIL Fab, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3W
 
 | | Cryo-EM structure of the chemokine-like receptor 1 in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3Y
 
 | | Cryo-EM structure of the G-protein coupled receptor 1 (GPR1) in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
1UEF
 
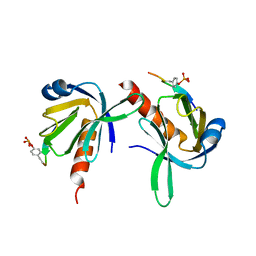 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
6KV5
 
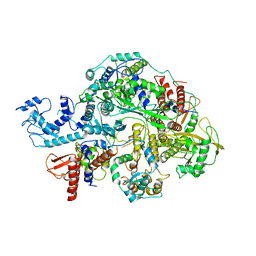 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
5D8W
 
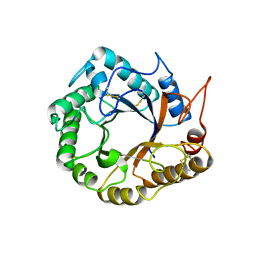 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
6KUJ
 
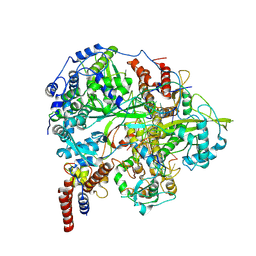 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 1 | | Descriptor: | 3'-cRNA promoter, 5'-cRNA promoter, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of influenza D virus polymerase bound to cRNA promoter in Mode A conformation
NAT NANOTECHNOL, 2019
|
|
4WIV
 
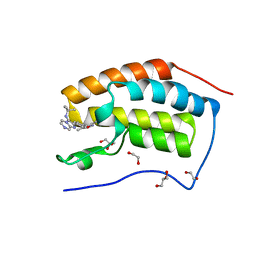 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a novel inhibitor UMB32 (N-TERT-BUTYL-2-[4-(3,5-DIMETHYL-1,2-OXAZOL-4-YL) PHENYL]IMIDAZO[1,2-A]PYRAZIN-3-AMINE) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, X, Blacklow, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biased multicomponent reactions to develop novel bromodomain inhibitors.
J.Med.Chem., 57, 2014
|
|
5DBF
 
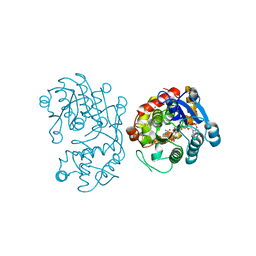 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NADPH | | Descriptor: | Iridoid synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DBI
 
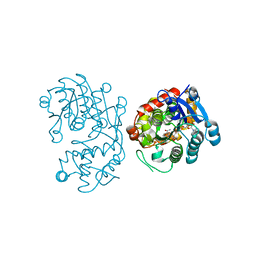 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ and 10-oxogeranial | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6TI2
 
 | |
