8KGM
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGP
 
 | |
8KGQ
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
7JNT
 
 | |
7Y0I
 
 | | Solution structures of ASH1L PHD domain in complex with H3K4me2 peptide | | Descriptor: | ALA-ARG-THR-MLY-GLN-THR-ALA-ARG-LYS-SER-THR-GLY-GLY-LYS-ALA, Histone-lysine N-methyltransferase ASH1L, ZINC ION | | Authors: | Yu, M, Zeng, L. | | Deposit date: | 2022-06-05 | | Release date: | 2022-10-12 | | Method: | SOLUTION NMR | | Cite: | Structural insight into ASH1L PHD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer.
Front Oncol, 12, 2022
|
|
7JY5
 
 | | Structure of human p97 in complex with ATPgammaS and Npl4/Ufd1 (masked around p97) | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative.
Nat Commun, 12, 2021
|
|
7YHQ
 
 | |
7YHP
 
 | |
8AGF
 
 | |
7K93
 
 | |
4WXI
 
 | |
7CVQ
 
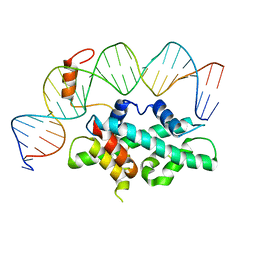 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
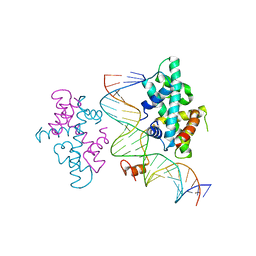 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
6HL8
 
 | |
6VAJ
 
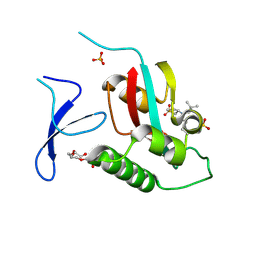 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
1F4J
 
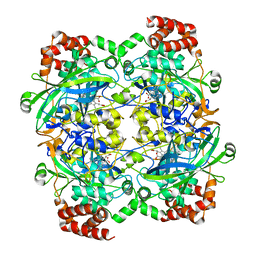 | | STRUCTURE OF TETRAGONAL CRYSTALS OF HUMAN ERYTHROCYTE CATALASE | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Safo, M.K, Musayev, F.N, Wu, S.H, Abraham, D.J, Ko, T.P. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of tetragonal crystals of human erythrocyte catalase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6W2D
 
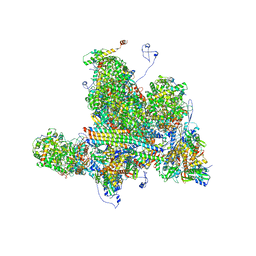 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
6W19
 
 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
6W2E
 
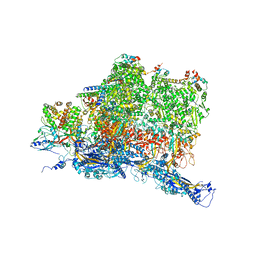 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
6KKI
 
 | |
6HL9
 
 | |
6KKL
 
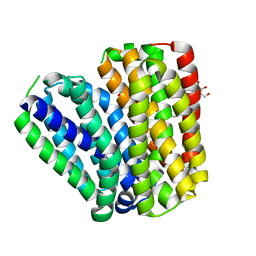 | | Crystal structure of Drug:Proton Antiporter-1 (DHA1) Family SotB, in the inward conformation (H115N mutant) | | Descriptor: | Sugar efflux transporter, nonyl beta-D-glucopyranoside | | Authors: | Xiao, Q.J, Sun, B, Zuo, Y.X, Guo, L, He, J.H, Deng, D. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Visualizing the nonlinear changes of a drug-proton antiporter from inward-open to occluded state.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6KKK
 
 | |
6KKJ
 
 | |
6KSW
 
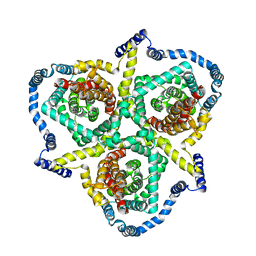 | | Cryo-EM structure of the human concentrative nucleoside transporter CNT3 | | Descriptor: | Solute carrier family 28 member 3 | | Authors: | Zhou, Y.X, Liao, L.H, Li, J.L, Xiao, Q.J, Sun, L.F, Deng, D. | | Deposit date: | 2019-08-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the human concentrative nucleoside transporter CNT3.
Plos Biol., 18, 2020
|
|
