7BEF
 
 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
3TFQ
 
 | | Crystal structure of 11b-hsd1 double mutant (l262r, f278e) complexed with 8-{[(2-CYANOPYRIDIN-3-YL)METHYL]SULFANYL}-6-HYDROXY-3,4-DIHYDRO-1H-PYRANO[3,4-C]PYRIDINE-5-CARBONITRILE | | Descriptor: | 8-{[(2-cyanopyridin-3-yl)methyl]sulfanyl}-6-hydroxy-3,4-dihydro-1H-pyrano[3,4-c]pyridine-5-carbonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 3-hydroxy-4-cyano-isoquinolines as novel, potent, and selective inhibitors of human 11beta-hydroxydehydrogenase 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3NJY
 
 | | Crystal structure of JMJD2A complexed with 5-carboxy-8-hydroxyquinoline | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | King, O.N.F, Clifton, I.J, Wang, M, Maloney, D.J, Jadhav, A, Oppermann, U, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Plos One, 5, 2010
|
|
7XXI
 
 | | Cryo-EM structure of the purinergic receptor P2Y12R in complex with 2MeSADP and Gi | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
7XXH
 
 | | Cryo-EM structure of the purinergic receptor P2Y1R in complex with 2MeSADP and G11 | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(11) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
7YNY
 
 | |
7YPO
 
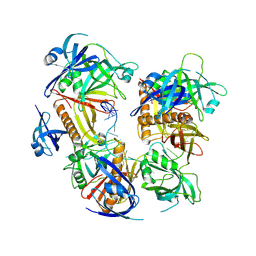 | | Cryo-EM structure of baculovirus LEF-3 in complex with ssDNA | | Descriptor: | DNA (28-MER), Lef3 | | Authors: | Yin, J, Fu, Y, Rao, G, Li, Z, Cao, S. | | Deposit date: | 2022-08-04 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA.
Nucleic Acids Res., 50, 2022
|
|
7YPQ
 
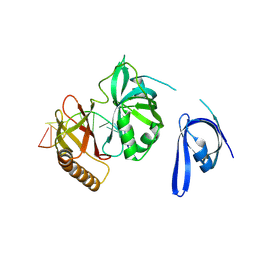 | | Cryo-EM structure of one baculovirus LEF-3 molecule in complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Lef3 | | Authors: | Fu, Y, Rao, G, Yin, J, Li, Z, Cao, S. | | Deposit date: | 2022-08-04 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA.
Nucleic Acids Res., 50, 2022
|
|
3PPT
 
 | | REP1-NXSQ fatty acid transporter | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-25 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7Y8W
 
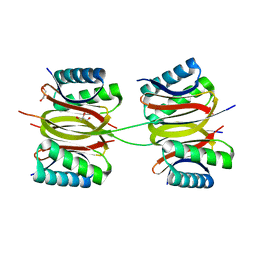 | | Crystal structure of DLC-1/SAO-1 complex | | Descriptor: | Dynein light chain 1, cytoplasmic, GLYCEROL, ... | | Authors: | Yan, H, Zhao, C, Wei, Z, Yu, C. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Cell Death Discov, 8, 2022
|
|
7Y61
 
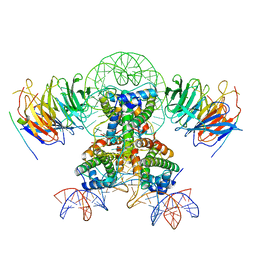 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5V
 
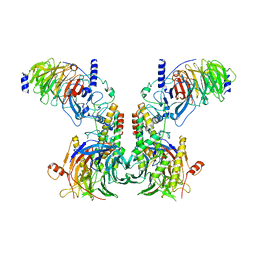 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y60
 
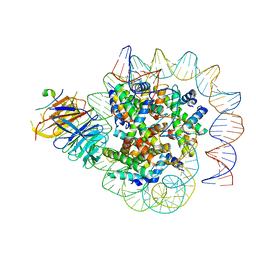 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5W
 
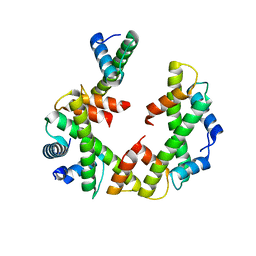 | | Cryo-EM structure of the left-handed Di-tetrasome | | Descriptor: | Histone H3.1, Histone H4, Widom 601 DNA (147-MER) | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5U
 
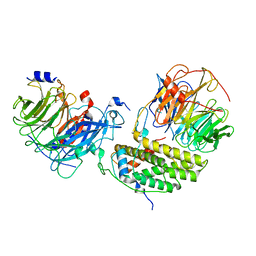 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
3PP6
 
 | | REP1-NXSQ fatty acid transporter Y128F mutant | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7YER
 
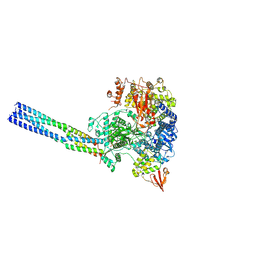 | | The structure of EBOV L-VP35 complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YET
 
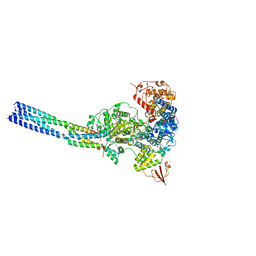 | | The structure of EBOV L-VP35 in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Polymerase cofactor VP35, RNA-directed RNA polymerase L | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YES
 
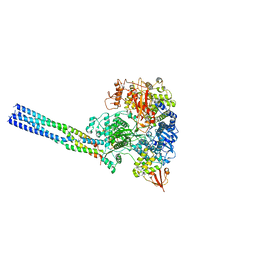 | |
7JS8
 
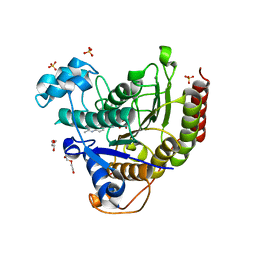 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH AN ETHYL KETONE INHIBITOR CONTAINING A SPIRO-BICYCLIC GROUP (COMPOUND 22) | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[4-(2-methylquinolin-6-yl)-1H-imidazol-2-yl]nonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity.
J.Med.Chem., 64, 2021
|
|
7JZV
 
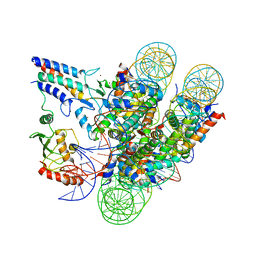 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6U9K
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 18 (TC-5153) | | Descriptor: | 5'-([(3S)-3-amino-3-carboxypropyl]{[1-(3,3-diphenylpropyl)azetidin-3-yl]methyl}amino)-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
5YBH
 
 | |
5YKF
 
 | |
6U9N
 
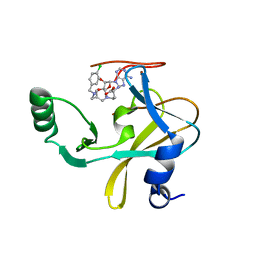 | | MLL1 SET N3861I/Q3867L bound to inhibitor 14 (TC-5139) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(4-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
