6V2H
 
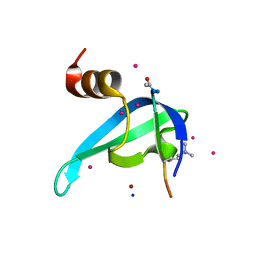 | | Crystal structure of CDYL2 in complex with H3tK27me3 | | Descriptor: | Chromodomain Y-like protein 2, H3tK27me3, NICKEL (II) ION, ... | | Authors: | Dong, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V41
 
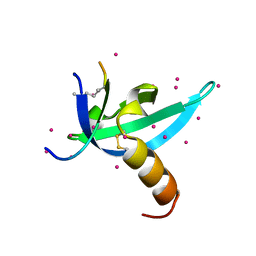 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V8W
 
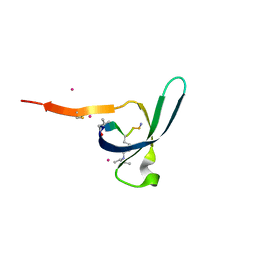 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V3N
 
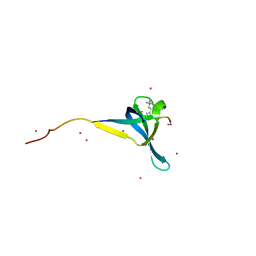 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
5HAD
 
 | |
6KGC
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 5.4 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGD
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 8.0 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
4DVF
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
6KGE
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 5.5 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KG8
 
 | | Solution structure of CaCohA2 from Clostridium acetobutylicum | | Descriptor: | Probably cellulosomal scaffolding protein, secreted cellulose-binding and cohesin domain | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
5I78
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I79
 
 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
8YOO
 
 | | Cryo-EM structure of the human 80S ribosome with 100 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8YOP
 
 | | Cryo-EM structure of the human 80S ribosome with 4 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSX
 
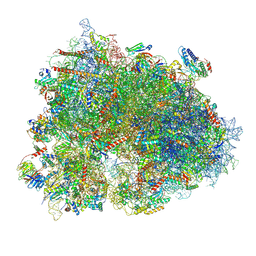 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA, SERBP1 and eEF2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSZ
 
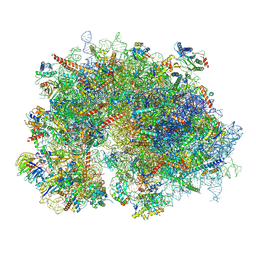 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA and P-tRNA | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSY
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, e-tRNA and CCDC124 (40S head Swivelled) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
5I77
 
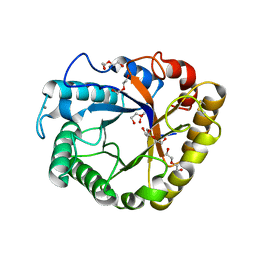 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
1C6X
 
 | |
1C6Y
 
 | |
4DV9
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-AMINO-3-OXOPROPYL)-2,13-DIBENZYL-12,22-DIHYDROXY-3,5,17-TRIMETHYL-8-(2-METHYLPROPYL)-4,7,10,15,18,21-HEXAOXO-19-(PROPAN-2-YL)-3,6,9,14,17,20-HEXAAZATRICOSAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE (NON-PREFERRED NAME), SULFATE ION | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
