6BW3
 
 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
1XX7
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001 | | Descriptor: | NICKEL (II) ION, UNKNOWN ATOM OR ION, oxetanocin-like protein | | Authors: | Chen, L, Tempel, W, Habel, J, Zhou, W, Nguyen, D, Chang, S.-H, Lee, D, Kelley, L.-L.C, Dillard, B.D, Liu, Z.-J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001
To be published
|
|
6C3S
 
 | |
1UX9
 
 | | Mapping protein matrix cavities in human cytoglobin through Xe atom binding: a crystallographic investigation | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | De Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping Protein Matrix Cavities in Human Cytoglobin Through Xe Atom Binding
Biochem.Biophys.Res.Commun., 316, 2004
|
|
6BWW
 
 | |
1Y02
 
 | | Crystal Structure of a FYVE-type domain from caspase regulator CARP2 | | Descriptor: | FYVE-RING finger protein SAKURA, ZINC ION | | Authors: | Tibbetts, M.D, Gu, L, Shiozaki, E.N, Shi, Y. | | Deposit date: | 2004-11-14 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a FYVE-type zinc finger domain from the caspase regulator CARP2.
STRUCTURE, 12, 2004
|
|
6J09
 
 | | Crystal structure of Haemophilus Influenzae BamA POTRA1-4 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | Deposit date: | 2018-12-21 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6D63
 
 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | Descriptor: | 3-oxopentanedioic acid, atzH | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | Deposit date: | 2018-04-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
1XK1
 
 | | Crystal Structures of the G139A, G139A-NO and G143H Mutants of Human Heme Oxygenase-1 | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lad, L, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2004-09-26 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of the G139A, G139A-NO and G143H mutants of human heme oxygenase-1. A finely tuned hydrogen-bonding network controls oxygenase versus peroxidase activity.
J.Biol.Inorg.Chem., 10, 2005
|
|
1UU4
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH CELLOBIOSE | | Descriptor: | ENDO-BETA-1,4-GLUCANASE, TETRAETHYLENE GLYCOL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
5ETA
 
 | | Structure of MAPK14 with bound the KIM domain of the Toxoplasma protein GRA24 | | Descriptor: | Mitogen-activated protein kinase 14, Putative transmembrane protein | | Authors: | Pellegrini, E, Palencia, A, Braun, L, Kapp, U, Bougdour, A, Belrhali, H, Bowler, M.W, Hakimi, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-10-26 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Subversion of MAP Kinase Signaling by an Intrinsically Disordered Parasite Secreted Agonist.
Structure, 25, 2017
|
|
1XUP
 
 | | ENTEROCOCCUS CASSELIFLAVUS GLYCEROL KINASE COMPLEXED WITH GLYCEROL | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Yeh, J.I, Charrier, V, Paulo, J, Hou, L, Darbon, E, Hol, W.G.J, Deutscher, J. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Enterococcal Glycerol Kinase in the Absence and Presence of Glycerol: Correlation of Conformation to Substrate Binding and a Mechanism of Activation by Phosphorylation
Biochemistry, 43, 2004
|
|
6DWK
 
 | | SAMHD1 Bound to Fludarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-fluoro-9-{5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-a mine, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1RR8
 
 | | Structural Mechanisms of Camptothecin Resistance by Mutations in Human Topoisomerase I | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T*GP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Chrencik, J.E, Staker, B.L, Burgin, A.B, Stewart, L, Redinbo, M.R. | | Deposit date: | 2003-12-08 | | Release date: | 2004-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of camptothecin resistance by human topoisomerase I mutations
J.Mol.Biol., 339, 2004
|
|
1XNV
 
 | | Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB), apo form #1 | | Descriptor: | propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
5EUM
 
 | | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae. | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipid A export ATP-binding/permease protein MsbA, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae.
To Be Published
|
|
1XR0
 
 | | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors | | Descriptor: | Basic fibroblast growth factor receptor 1, FGFR signalling adaptor SNT-1 | | Authors: | Dhalluin, C, Yan, K.S, Plotnikova, O, Lee, K.W, Zeng, L, Kuti, M, Mujtaba, S, Goldfarb, M.P, Zhou, M.-M. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors
Mol.Cell, 6, 2000
|
|
5EUL
 
 | | Structure of the SecA-SecY complex with a translocating polypeptide substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AYC08, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Li, L, Park, E, Ling, J, Ingram, J, Ploegh, H, Rapoport, T.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of a substrate-engaged SecY protein-translocation channel.
Nature, 531, 2016
|
|
5F0G
 
 | |
2LAL
 
 | | CRYSTAL STRUCTURE DETERMINATION AND REFINEMENT AT 2.3 ANGSTROMS RESOLUTION OF THE LENTIL LECTIN | | Descriptor: | CALCIUM ION, LENTIL LECTIN (ALPHA CHAIN), LENTIL LECTIN (BETA CHAIN), ... | | Authors: | Loris, R, Steyaert, J, Maes, D, Lisgarten, J, Pickersgill, R, Wyns, L. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
5EVG
 
 | | Crystal structure of a Francisella virulence factor FvfA in the orthorhombic form | | Descriptor: | Francisella virulence factor | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1XQZ
 
 | | Crystal Structure of hPim-1 kinase at 2.1 A resolution | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Qian, K.C, Wang, L, Hickey, E.R, Studts, J, Barringer, K, Peng, C, Kronkaitis, A, Li, J, White, A, Mische, S, Farmer, B. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Constitutive Activity and a Unique Nucleotide Binding Mode of Human Pim-1 Kinase.
J.Biol.Chem., 280, 2005
|
|
5CTG
 
 | | The 3.1 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Tao, Y, Perry, K, Feng, L. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
5CTH
 
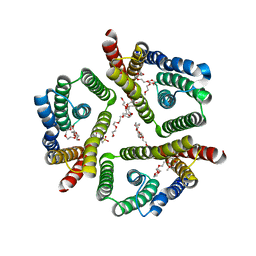 | | The 3.7 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Feng, L, Tao, Y, Perry, K. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
1XSQ
 
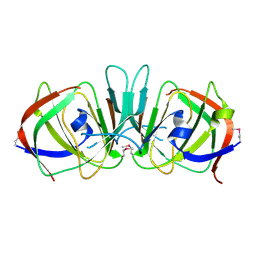 | | Crystal structure of ureidoglycolate hydrolase from E.coli. Northeast Structural Genomics Consortium target ET81. | | Descriptor: | Ureidoglycolate hydrolase | | Authors: | Kuzin, A.P, Vorobiev, S.M, Abashidze, M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase from E.coli. Northeast Structural Genomics Consortium target ET81.
To be Published
|
|
