1V5P
 
 | | Solution Structure of the N-terminal Pleckstrin Homology Domain Of TAPP2 from Mouse | | Descriptor: | pleckstrin homology domain-containing, family A | | Authors: | Li, H, Hayashi, F, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Pleckstrin Homology Domain Of TAPP2 from Mouse
To be Published
|
|
1V70
 
 | |
1V8Z
 
 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
1V9X
 
 | |
1V25
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, long-chain-fatty-acid-CoA synthetase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1V3U
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase in apo form | | Descriptor: | CHLORIDE ION, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1V6B
 
 | | Solution structure of the third PDZ domain of mouse harmonin | | Descriptor: | harmonin isoform a1 | | Authors: | Yamada, K, Nameki, N, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-28 | | Release date: | 2004-05-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third PDZ domain of mouse harmonin
To be Published
|
|
2RPC
 
 | |
2RQC
 
 | | Solution Structure of RNA-binding domain 3 of CUGBP1 in complex with RNA (UG)3 | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*G)-3', CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 37, 2009
|
|
2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
2PA2
 
 | | Crystal structure of human Ribosomal protein L10 core domain | | Descriptor: | 60S ribosomal protein L10, POTASSIUM ION | | Authors: | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
2P6C
 
 | |
2PMH
 
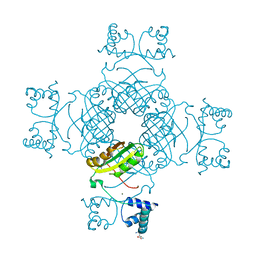 | | Crystal structure of Thr132Ala of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-22 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
1UKK
 
 | | Structure of Osmotically Inducible Protein C from Thermus thermophilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Osmotically Inducible Protein C | | Authors: | Rehse, P.H, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Structure and Biochemical Analysis of the Thermus thermophilus Osmotically Inducible Protein C
J.MOL.BIOL., 338, 2004
|
|
1UK5
 
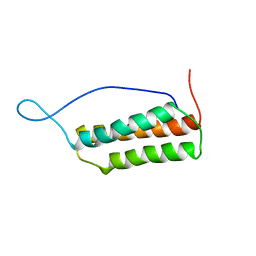 | | Solution structure of the Murine BAG domain of Bcl2-associated athanogene 3 | | Descriptor: | BAG-family molecular chaperone regulator-3 | | Authors: | Hatta, R, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-19 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
1V5J
 
 | | Solution Structure of RSGI RUH-008, fn3 domain in Human cDNA | | Descriptor: | KIAA1355 protein | | Authors: | Doi-Katayama, Y, Hayashi, F, Hirota, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-008, a fn3 domain in Human cDNA
To be Published
|
|
1V2B
 
 | | Crystal Structure of PsbP Protein in the Oxygen-Evolving Complex of Photosystem II from Higher Plants | | Descriptor: | 23-kDa polypeptide of photosystem II oxygen-evolving complex, SULFATE ION, alpha-D-glucopyranose | | Authors: | Ifuku, K, Nakatsu, T, Kato, H, Sato, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-14 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum
Embo Rep., 5, 2004
|
|
1VCG
 
 | | Crystal Structure of IPP isomerase at P43212 | | Descriptor: | FLAVIN MONONUCLEOTIDE, isopentenyl-diphosphate delta-isomerase | | Authors: | Wada, T, Park, S.-Y, Tame, R.H, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal Structure of IPP isomerase at P43212
To be Published
|
|
1V33
 
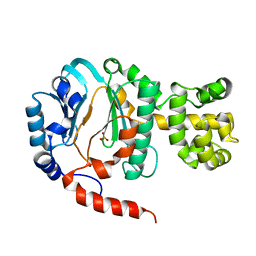 | | Crystal structure of DNA primase from Pyrococcus horikoshii | | Descriptor: | DNA primase small subunit, PHOSPHATE ION, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1V85
 
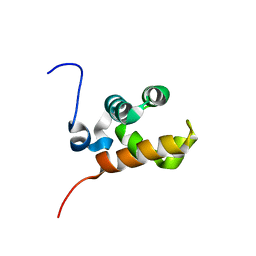 | | Sterile alpha motif (SAM) domain of mouse bifunctional apoptosis regulator | | Descriptor: | similar to ring finger protein 36 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Hayashi, F, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sterile alpha motif (SAM) domain of mouse bifunctional apoptosis regulator
To be Published
|
|
1V34
 
 | | Crystal structure of Pyrococcus horikoshii DNA primase-UTP complex | | Descriptor: | DNA primase small subunit, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1V9A
 
 | | Crystal structure of Uroporphyrin-III C-methyl transferase from Thermus thermophilus complexed with S-adenyl homocysteine | | Descriptor: | CITRATE ANION, S-ADENOSYL-L-HOMOCYSTEINE, Uroporphyrin-III C-methyltransferase | | Authors: | Rehse, P.H, Kitao, T, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-23 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1VA0
 
 | | Crystal Structure of the Native Form of Uroporphyrin III C-methyl transferase from Thermus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Uroporphyrin-III C-methyltransferase | | Authors: | Rehse, P.H, Kitao, T, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WW8
 
 | |
1X4C
 
 | | Solution structure of RRM domain in splicing factor 2 | | Descriptor: | Splicing factor, arginine/serine-rich 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in splicing factor 2
To be Published
|
|
