1MDX
 
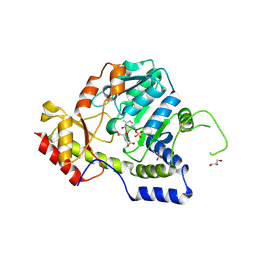 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ArnB aminotransferase, GLYCEROL, ... | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
3BO8
 
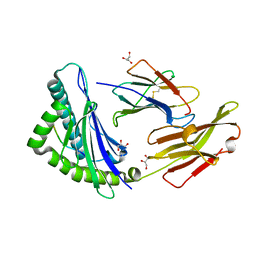 | | The High Resolution Crystal Structure of HLA-A1 Complexed with the MAGE-A1 Peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational changes within the HLA-A1:MAGE-A1 complex induced by binding of a recombinant antibody fragment with TCR-like specificity
Protein Sci., 18, 2009
|
|
2POX
 
 | |
1ARJ
 
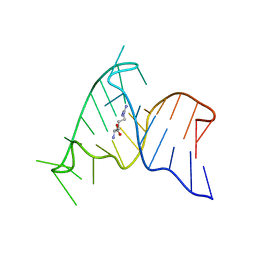 | | ARG-BOUND TAR RNA, NMR | | Descriptor: | ARGININE, TAR RNA | | Authors: | Aboul-Ela, F, Varani, G, Karn, J. | | Deposit date: | 1995-08-30 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein.
J.Mol.Biol., 253, 1995
|
|
1B9Q
 
 | |
2Q2C
 
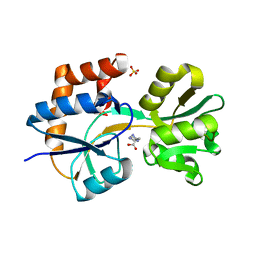 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, GLYCEROL, HISTIDINE, ... | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2PVU
 
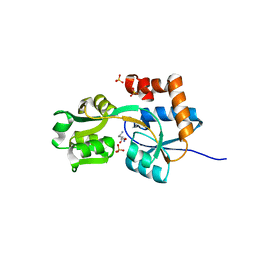 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, LYSINE, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
1YGW
 
 | |
1B9P
 
 | |
1AUW
 
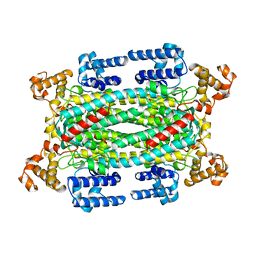 | | H91N DELTA 2 CRYSTALLIN FROM DUCK | | Descriptor: | DELTA 2 CRYSTALLIN | | Authors: | Abu-Abed, M, Vallee, F, Howell, P.L. | | Deposit date: | 1997-09-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of the enzymatically active and inactive forms of delta crystallin and the role of histidine 91.
Biochemistry, 36, 1997
|
|
2Q2A
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ARGININE, ArtJ, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-26 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
7ZBN
 
 | |
7Z53
 
 | |
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
7YOT
 
 | |
2LEV
 
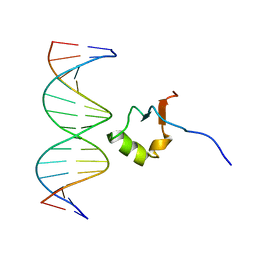 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
2QYP
 
 | |
2R0R
 
 | |
1ANR
 
 | |
2R1Q
 
 | |
1INN
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, P21 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
2RB3
 
 | | Crystal Structure of Human Saposin D | | Descriptor: | GLYCEROL, Proactivator polypeptide, SULFATE ION | | Authors: | Maier, T, Rossman, M, Saenger, W. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human saposins C and d: implications for lipid recognition and membrane interactions.
Structure, 16, 2008
|
|
2ISC
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with DADMe-Imm-A | | Descriptor: | (3R,4R)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-(HYDROXYMETHYL)PYRROLIDIN-3-OL, PHOSPHATE ION, purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2006-10-17 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues
Biochemistry, 46, 2007
|
|
5NHW
 
 | | CRYSTAL STRUCTURE OF THE BIMAGRUMAB Fab | | Descriptor: | GLYCEROL, anti-human ActRII Bimagrumab Fab heavy-chain, anti-human ActRII Bimagrumab Fab light-chain | | Authors: | Rondeau, J.-M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Blockade of activin type II receptors with a dual anti-ActRIIA/IIB antibody is critical to promote maximal skeletal muscle hypertrophy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7X3X
 
 | | Cryo-EM structure of N1 nucleosome-RA | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
