3H0F
 
 | | Crystal structure of the human Fyn SH3 R96W mutant | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Ponchon, L, Hoh, F, Labesse, G, Dumas, C, Arold, S.T. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
7CYP
 
 | | Complex of SARS-CoV-2 spike trimer with its neutralizing antibody HB27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of HB27, ... | | Authors: | Wang, X, Zhu, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-06-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
4ANW
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-6-{4-CHLORO-3-[(2,3-DIFLUOROPHENYL)SULFAMOYL]PHENYL}-N-METHYLPYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4DJV
 
 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DJY
 
 | |
3H0H
 
 | |
5FI7
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL_00015: 2-phenyl-~{N}-[5-[(3~{S})-3-[[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]oxy]pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl]ethanamide | | Descriptor: | 2-phenyl-~{N}-[5-[(3~{S})-3-[[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]oxy]pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
8W41
 
 | | Cryo-EM structure of Myosin VI in the autoinhibited state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin-1, ... | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Autoinhibition and activation of myosin VI revealed by its cryo-EM structure.
Nat Commun, 15, 2024
|
|
5U62
 
 | | Crystal structure of EED in complex with H3K27Me3 peptide and 6-(benzo[d][1,3]dioxol-4-ylmethyl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2H-1,3-benzodioxol-4-yl)methyl]-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
6J07
 
 | | Crystal structure of human TERB2 and TERB1 | | Descriptor: | Telomere repeats-binding bouquet formation protein 1, Telomere repeats-binding bouquet formation protein 2, ZINC ION | | Authors: | Wang, Y, Chen, Y, Wu, J, Huang, C, Lei, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | The meiotic TERB1-TERB2-MAJIN complex tethers telomeres to the nuclear envelope.
Nat Commun, 10, 2019
|
|
5U5K
 
 | |
4F1S
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine-sulfonamide inhibitor | | Descriptor: | N-(5-{[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-5-(tetrahydro-2H-pyran-4-yl)pyridin-2-yl]amino}-2-chloropyridin-3-yl)methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationships of dual PI3K/mTOR inhibitors based on a 4-amino-6-methyl-1,3,5-triazine sulfonamide scaffold.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5GNK
 
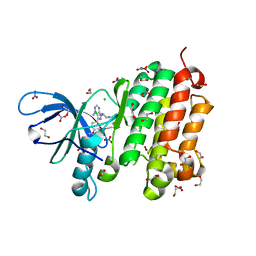 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
4GE6
 
 | |
5GTY
 
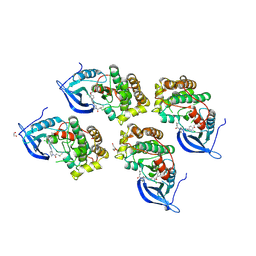 | | Crystal structure of EGFR 696-1022 T790M in complex with LXX-6-26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(6-methylpyridin-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode.
Oncotarget, 8, 2017
|
|
5U5T
 
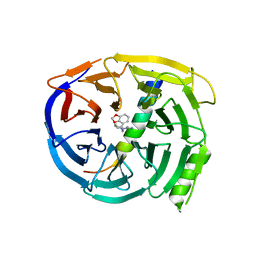 | | Crystal structure of EED in complex with H3K27Me3 peptide and 3-(benzo[d][1,3]dioxol-4-ylmethyl)piperidine-1-carboximidamide | | Descriptor: | (3R)-3-[(2H-1,3-benzodioxol-4-yl)methyl]piperidine-1-carboximidamide, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
4GE2
 
 | |
5FI2
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL_00009: 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol- 2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
4GE5
 
 | |
5U5H
 
 | | Crystal structure of EED in complex with 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2-fluoro-5-methoxyphenyl)methyl]-1-(propan-2-yl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Polycomb protein EED | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
3N2Y
 
 | |
7SUS
 
 | | Crystal structure of Apelin receptor in complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Han, G.W, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5FI6
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL_00011: 2-phenyl-~{N}-[5-[[(3~{S})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{S})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
7CWN
 
 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
8YFN
 
 | |
