3WWF
 
 | | Crystal structure of the computationally designed Pizza2-SR protein | | Descriptor: | pizza2sr-pb protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WWB
 
 | | Crystal structure of the computationally designed Pizza2-SR protein | | Descriptor: | Pizza2-SR protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WWA
 
 | | Crystal structure of the computationally designed Pizza7 protein after heat treatment | | Descriptor: | ISOPROPYL ALCOHOL, Pizza7H protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WW9
 
 | | Crystal structure of the computationally designed Pizza6 protein | | Descriptor: | GLYCEROL, SULFATE ION, pizza6 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WW7
 
 | | Crystal structure of the computationally designed Pizza2 protein | | Descriptor: | GLYCEROL, Pizza2 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8XM7
 
 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex: RhoG/DOCK5/ELMO1 focused map | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-12-27 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.91 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJ2
 
 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8JHK
 
 | | Cryo-EM structure of the DOCK5/ELMO1 complex, focused on one protomer | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
7COZ
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-41 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(2-methyl-4-thiophen-2-yl-1,3-thiazol-5-yl)propanoyl]piperidine-4-carboxamide, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
7CP0
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
7DPA
 
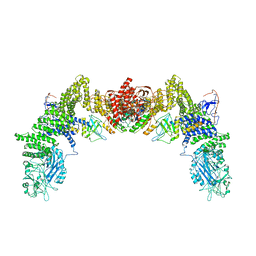 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
7D8E
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-09 | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-09
To Be Published
|
|
7DVH
 
 | |
7DVC
 
 | | Crystal structure of the computationally designed reDPBB_sym1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, reDPBB_sym1 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
7DVF
 
 | | Crystal structure of the computationally designed reDPBB_sym2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, reDPBB_sym2 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
4HVS
 
 | |
4HW7
 
 | |
4FK3
 
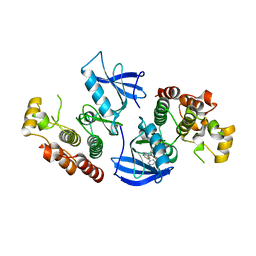 | | B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 | | Descriptor: | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Wang, W, Zhang, K.Y.J. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5F53
 
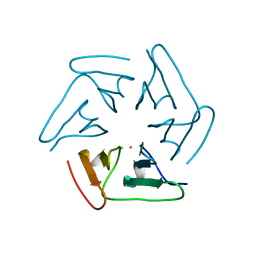 | |
5I1Z
 
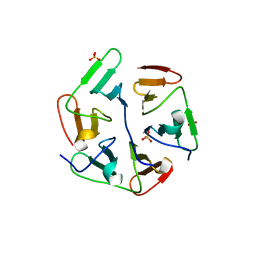 | | Structure of nvPizza2-H16S58 | | Descriptor: | SULFATE ION, nvPizza2-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broken Symmetry: Partial domain swapping in an artificial trimeric protein
To Be Published
|
|
6JPP
 
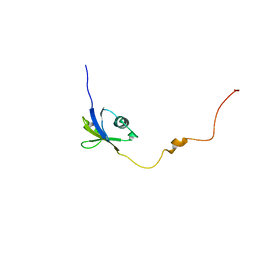 | |
6JMN
 
 | | Crystal structure of Ostrinia furnacalis Chitinase h complexed with compound 2-8-s2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitinase | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-03-12 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JJR
 
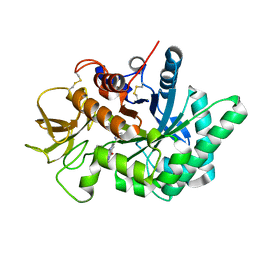 | | Crystal structure of human chitotriosidase-1 (hChit1) catalytic domain in complex with compound 2-8-14 | | Descriptor: | 6-azanyl-2-oxidanylidene-N-[(1S)-1-phenylethyl]-7-(phenylmethyl)-1$l^{4},9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-1(14),3(8),4,6,10,12-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JK9
 
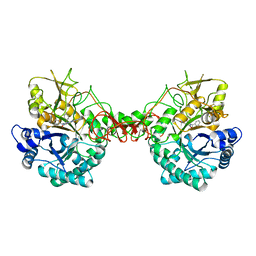 | | Crystal structure of Serratia marcescens Chitinase B complexed with compound 2-8-14 | | Descriptor: | 6-azanyl-2-oxidanylidene-N-[(1S)-1-phenylethyl]-7-(phenylmethyl)-1$l^{4},9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-1(14),3(8),4,6,10,12-hexaene-5-carboxamide, Chitinase | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-27 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JK6
 
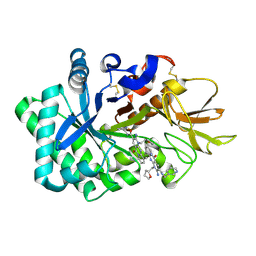 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 2-8-s2 | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-27 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
