7ULS
 
 | | Recombinant muscarinic toxin alpha | | Descriptor: | GLYCEROL, Muscarinic toxin alpha | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | recombinant MTalpha at 1.8 Angstroms resolution
To Be Published
|
|
7ULG
 
 | | recombinant alpha cobra toxin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Alpha-cobratoxin | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-04 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of recombinant alpha cobra toxin at 1.57 Angstroms
To Be Published
|
|
7ULQ
 
 | | Recombinant Hannalgesin | | Descriptor: | ACETATE ION, GLYCEROL, Long neurotoxin OH-55, ... | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant Hannalgesin at 2.2 Angstroms resolution.
To Be Published
|
|
7EJ0
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJA
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to dexmedetomidine | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJ8
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to brimonidine | | Descriptor: | Alpha-2A adrenergic receptor, Brimonidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJK
 
 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to oxymetazoline | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7DRX
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-30 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSH
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSI
 
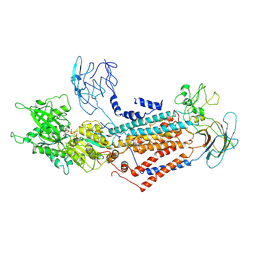 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with AMPPCP ( resting state ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7WHV
 
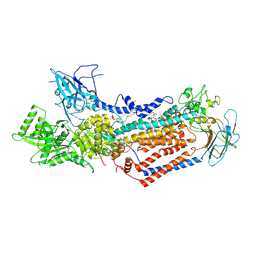 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7WHW
 
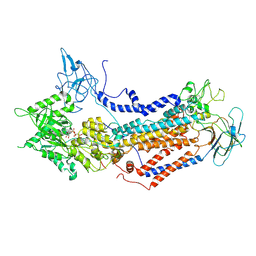 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7F7F
 
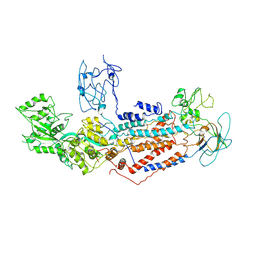 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with beryllium fluoride (resting state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-06-29 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
5XE1
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with INCB14943 | | Descriptor: | 4-Amino-N-(3-chloro-4-fluorophenyl)-N'-hydroxy-1,2,5-oxadiazole-3-carboxamidine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, J, Wu, U, Liu, J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the binding mechanism of IDO1 with hydroxylamidine based inhibitor INCB14943
Biochem. Biophys. Res. Commun., 487, 2017
|
|
4NPK
 
 | | Extended-Synaptotagmin 2, C2A- and C2B-domains, calcium bound | | Descriptor: | CALCIUM ION, Extended synaptotagmin-2 | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2013-11-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Structure and ca(2+)-binding properties of the tandem c2 domains of e-syt2.
Structure, 22, 2014
|
|
4ZRD
 
 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4ZRE
 
 | | Crystal structure of SMG1 F278D mutant | | Descriptor: | CHLORIDE ION, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
8HRJ
 
 | |
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRK
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 complex | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRN
 
 | | Cryo-EM structure of ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRU
 
 | |
8HRI
 
 | |
8HRM
 
 | | Cryo-EM structure of streptavidin | | Descriptor: | Streptavidin | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
