4KXY
 
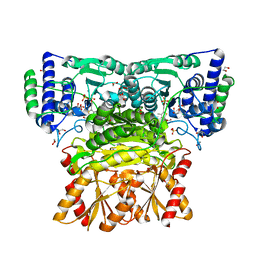 | | Human transketolase in complex with ThDP analogue (R)-2-(1,2-dihydroxyethyl)-3-deaza-ThDP | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-[(1R)-1,2-dihydroxyethyl]-3-methylthiophen-2-yl}ethyl trihydrogen diphosphate, CALCIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Erixon, K.M, Leeper, F, Kluger, R, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXW
 
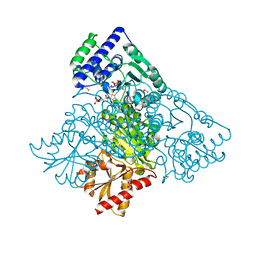 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 2 | | Descriptor: | 1,2-ETHANEDIOL, D-XYLITOL-5-PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
3CGN
 
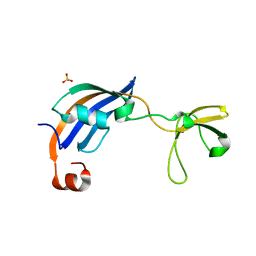 | | Crystal Structure of thermophilic SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Neumann, P, Loew, C, Stubbs, M.T, Balbach, J. | | Deposit date: | 2008-03-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
3FT7
 
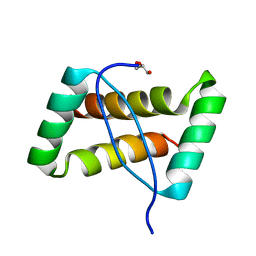 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | Descriptor: | GLYCEROL, Uncharacterized protein ORF56 | | Authors: | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | Deposit date: | 2009-01-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
8ARU
 
 | |
5L4W
 
 | | Crystal structure of FimH lectin domain in complex with 3-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4U
 
 | | Crystal structure of FimH lectin domain in complex with 2-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4Y
 
 | | Crystal structure of FimH lectin domain in complex with 4-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4X
 
 | | Crystal structure of FimH lectin domain in complex with 4-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4V
 
 | | Crystal structure of FimH lectin domain in complex with 3-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4T
 
 | | Crystal structure of FimH lectin domain in complex with 2-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5HSA
 
 | | Alcohol Oxidase AOX1 from Pichia Pastoris | | Descriptor: | ARABINO-FLAVIN-ADENINE DINUCLEOTIDE, Alcohol oxidase 1, CALCIUM ION, ... | | Authors: | Neumann, P, Ficner, R, Feussner, I, Koch, C. | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alcohol Oxidase from Pichia pastoris.
Plos One, 11, 2016
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2VQW
 
 | | Structure of inhibitor-free HDAC4 catalytic domain (with gain-of- function mutation His332Tyr) | | Descriptor: | HISTONE DEACETYLASE 4, POTASSIUM ION, ZINC ION | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQM
 
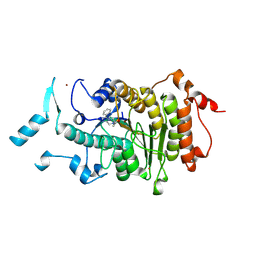 | | Structure of HDAC4 catalytic domain bound to a hydroxamic acid inhbitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
3E7X
 
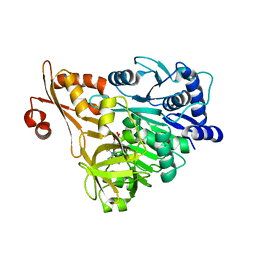 | | Crystal structure of DLTA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1 | | Authors: | Yonus, H, Neumann, P, Zimmermann, S, May, J.J, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains
J.Biol.Chem., 283, 2008
|
|
3EY6
 
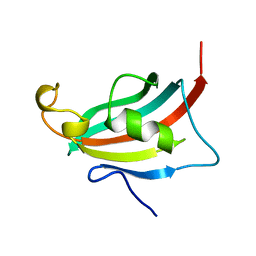 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
5EQG
 
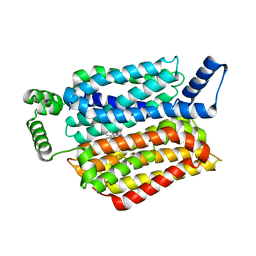 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EK8
 
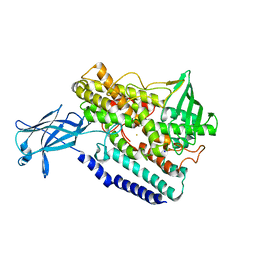 | | Crystal structure of a 9R-lipoxygenase from Cyanothece PCC8801 at 2.7 Angstroms | | Descriptor: | FE (II) ION, Lipoxygenase, SODIUM ION | | Authors: | Feussner, I, Ficner, R, Neumann, P, Newie, J, Andreou, A, Einsle, O. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a lipoxygenase from Cyanothece sp. may reveal novel features for substrate acquisition.
J.Lipid Res., 57, 2016
|
|
5EQI
 
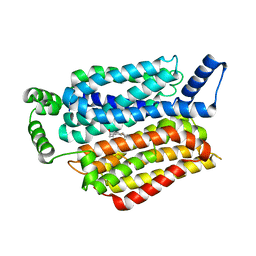 | | Human GLUT1 in complex with Cytochalasin B | | Descriptor: | Cytochalasin B, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EQH
 
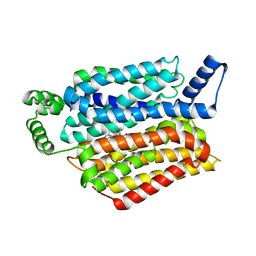 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3NC0
 
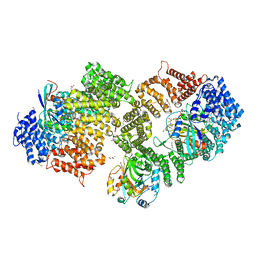 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal II) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exportin-1, GLYCEROL, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NBY
 
 | | Crystal structure of the PKI NES-CRM1-RanGTP nuclear export complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NBZ
 
 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal I) | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NC1
 
 | | Crystal structure of the CRM1-RanGTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
