4NWL
 
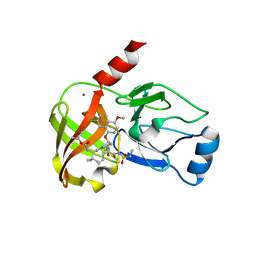 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | Descriptor: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4IBF
 
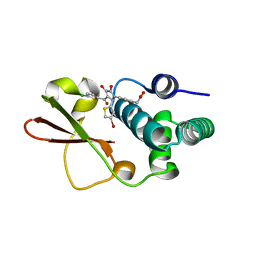 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBE
 
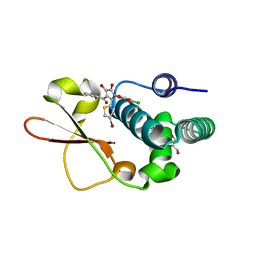 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-chlorobenzoic acid, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBK
 
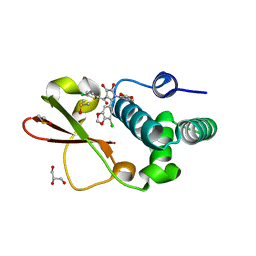 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBC
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | DIMETHYL SULFOXIDE, Polymerase cofactor VP35, {4-[(2R)-3-(2-chlorobenzoyl)-2-(2-chlorophenyl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4NWK
 
 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4IBG
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
5EZS
 
 | |
5EZQ
 
 | |
3WDZ
 
 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | Descriptor: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
7L6M
 
 | |
7LUA
 
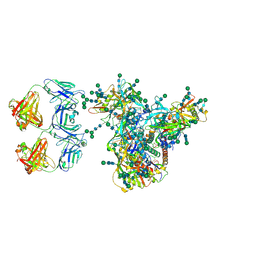 | | Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 SOSIP gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7LU9
 
 | | Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-04-14 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
1G6Q
 
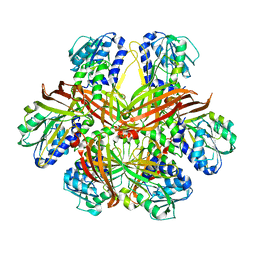 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
2EZZ
 
 | |
2GKD
 
 | |
2EZX
 
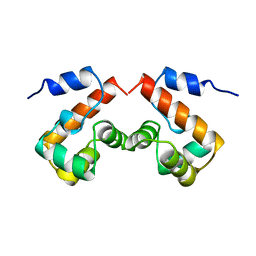 | |
2EZY
 
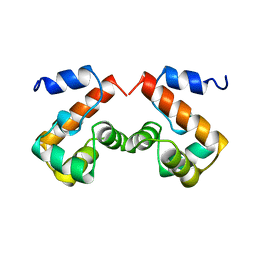 | |
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
7JVV
 
 | | Crystal structure of human histone deacetylase 8 (HDAC8) E66D/Y306F double mutation complexed with a tetrapeptide substrate | | Descriptor: | 1,2-ETHANEDIOL, ACE-ARG-HIS-ALY-ALY-MCM, GLYCEROL, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7JVU
 
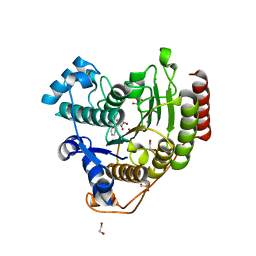 | | Crystal structure of human histone deacetylase 8 (HDAC8) I45T mutation complexed with SAHA | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5004766 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
7JVW
 
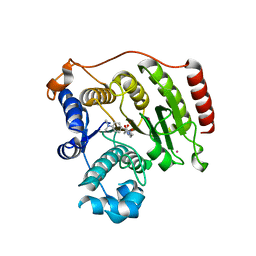 | | Crystal structure of human histone deacetylase 8 (HDAC8) G320R mutation complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W, Decroos, C, Porter, N.J, Lee, M. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40301776 Å) | | Cite: | Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
J.Struct.Biol., 213, 2020
|
|
2Q44
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At1g77540 | | Descriptor: | BROMIDE ION, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
