1DHR
 
 | | CRYSTAL STRUCTURE OF RAT LIVER DIHYDROPTERIDINE REDUCTASE | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Skinner, M.M, Whiteley, J.M, Matthews, D.A, Xuong, N.H. | | Deposit date: | 1992-03-30 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rat liver dihydropteridine reductase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1DKF
 
 | | CRYSTAL STRUCTURE OF A HETERODIMERIC COMPLEX OF RAR AND RXR LIGAND-BINDING DOMAINS | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, OLEIC ACID, PROTEIN (RETINOIC ACID RECEPTOR-ALPHA), ... | | Authors: | Bourguet, W, Vivat, V, Wurtz, J.M, Chambon, P, Gronemeyer, H, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1999-12-07 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains.
Mol.Cell, 5, 2000
|
|
1DI1
 
 | | CRYSTAL STRUCTURE OF ARISTOLOCHENE SYNTHASE FROM PENICILLIUM ROQUEFORTI | | Descriptor: | ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W, Rynkiewicz, M.J. | | Deposit date: | 1999-11-28 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
1DDK
 
 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
4R27
 
 | | Crystal structure of beta-glycosidase BGL167 | | Descriptor: | Glycoside hydrolase | | Authors: | Park, S.J, Choi, J.M, Kyeong, H.H, Kim, S.G, Kim, H.S. | | Deposit date: | 2014-08-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational design of a beta-glycosidase with high regiospecificity for triterpenoid tailoring
Chembiochem, 16, 2015
|
|
1DFP
 
 | | FACTOR D INHIBITED BY DIISOPROPYL FLUOROPHOSPHATE | | Descriptor: | DIISOPROPYL PHOSPHONATE, FACTOR D | | Authors: | Cole, L.B, Chu, N, Kilpatrick, J.M, Volanakis, J.E, Narayana, S.V.L, Babu, Y.S. | | Deposit date: | 1997-02-18 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of diisopropyl fluorophosphate-inhibited factor D.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
4BA7
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Bacteria Common Ancestor (LBCA) from the Precambrian Period | | Descriptor: | 1,2-ETHANEDIOL, LBCA THIOREDOXIN | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-09-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
1DG2
 
 | | SOLUTION CONFORMATION OF A-CONOTOXIN AUIB | | Descriptor: | A-CONOTOXIN AUIB | | Authors: | Cho, J.-H, Mok, K.H, Olivera, B.M, McIntosh, J.M, Park, K.-H, Han, K.-H. | | Deposit date: | 1999-11-23 | | Release date: | 2000-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution conformation of alpha-conotoxin AuIB, an alpha(3)beta(4) subtype-selective neuronal nicotinic acetylcholine receptor antagonist.
J.Biol.Chem., 275, 2000
|
|
1DL8
 
 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
4R7Z
 
 | | PfMCM-AAA double-octamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 21, MAGNESIUM ION | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
1DD6
 
 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DB1
 
 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | Authors: | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
3G7C
 
 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
1DD9
 
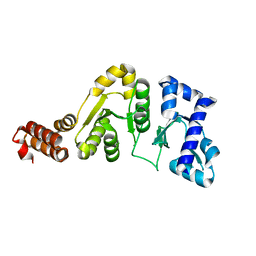 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | Descriptor: | DNA PRIMASE, STRONTIUM ION | | Authors: | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | Deposit date: | 1999-11-09 | | Release date: | 2000-04-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1HB2
 
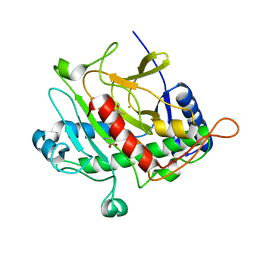 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN EXPOSED PRODUCT FROM ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1S)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOCARBONYL)-2-OXOETHYL]-6-OXO-L-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
1DCW
 
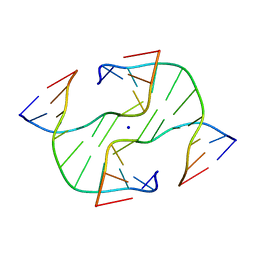 | | STRUCTURE OF A FOUR-WAY JUNCTION IN AN INVERTED REPEAT SEQUENCE. | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3'), SODIUM ION | | Authors: | Eichman, B.F, Vargason, J.M, Mooers, B.H.M, Ho, P.S. | | Deposit date: | 1999-11-05 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DDB
 
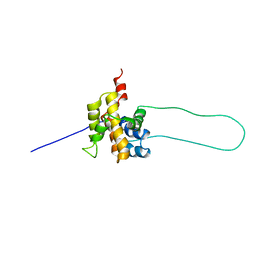 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (BID) | | Authors: | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | Deposit date: | 1999-02-19 | | Release date: | 1999-08-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
4R1P
 
 | |
4R7Y
 
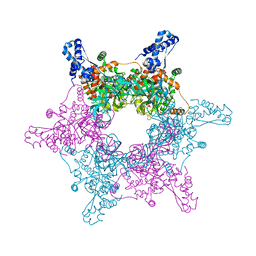 | | Crystal structure of an active MCM hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
1DGP
 
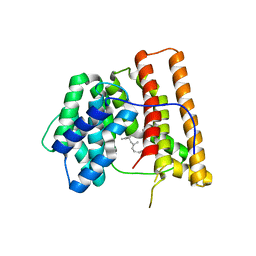 | | ARISTOLOCHENE SYNTHASE FARNESOL COMPLEX | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W. | | Deposit date: | 1999-11-24 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
1H96
 
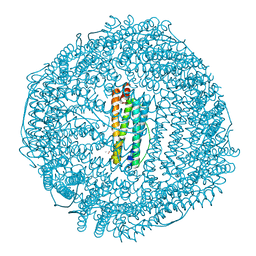 | | recombinant mouse L-chain ferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, SULFATE ION | | Authors: | Granier, T, Gallois, B, D'Estaintot, B.L, Dautant, A, Chevalier, J.M, Mellado, J.M, Beaumont, C, Santambrogio, P, Arosio, P, Precigoux, G. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mouse L-Chain Ferritin at 1.6 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4R6X
 
 | |
1DI2
 
 | |
1DKI
 
 | | CRYSTAL STRUCTURE OF THE ZYMOGEN FORM OF STREPTOCOCCAL PYROGENIC EXOTOXIN B ACTIVE SITE (C47S) MUTANT | | Descriptor: | PYROGENIC EXOTOXIN B ZYMOGEN, SULFATE ION | | Authors: | Kagawa, T.F, Cooney, J.C, Baker, H.M, McSweeney, S, Liu, M, Gubba, S, Musser, J.M, Baker, E.N. | | Deposit date: | 1999-12-07 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1HB3
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN EXPOSED PRODUCT FROM ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1S)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOCARBONYL)-2-OXOETHYL]-6-OXO-L-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
