3SKO
 
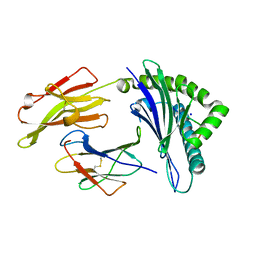 | | Crystal structure of the HLA-B8-A66-FLR, mutant A66 of the HLA B8 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
2Y6D
 
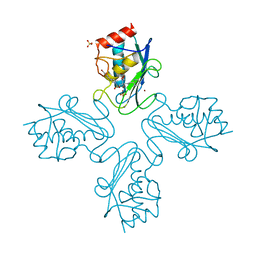 | | The Discovery of MMP7 Inhibitors Exploiting a Novel Selectivity Trigger | | Descriptor: | CALCIUM ION, MATRILYSIN, N-[(2S)-1-[4-(5-BROMOPYRIDIN-2-YL)PIPERAZIN-1-YL]SULFONYL-5-PYRIMIDIN-2-YL-PENTAN-2-YL]-N-HYDROXY-METHANAMIDE, ... | | Authors: | Edman, K, Furber, M, Hemsley, P, Johansson, C, Pairaudeau, G, Petersen, J, Stocks, M, Tervo, A, Ward, A, Wells, E, Wissler, L. | | Deposit date: | 2011-01-20 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Discovery of Mmp7 Inhibitors Exploiting a Novel Selectivity Trigger.
Chemmedchem, 6, 2011
|
|
2YB8
 
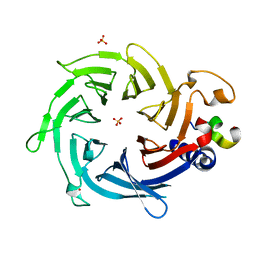 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
2X9E
 
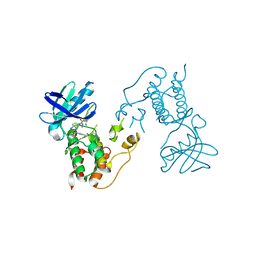 | | HUMAN MPS1 IN COMPLEX WITH NMS-P715 | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, N-(2,6-DIETHYLPHENYL)-1-METHYL-8-({4-[(1-METHYLPIPERIDIN-4-YL)CARBAMOYL]-2-(TRIFLUOROMETHOXY)PHENYL}AMINO)-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE | | Authors: | Colombo, R, Caldarelli, M, Mennecozzi, M, Giorgini, M.L, Sola, F, Cappella, P, Perrera, C, DePaolini, S.R, Rusconi, L, Cucchi, U, Avanzi, N, Bertrand, J.A, Bossi, R.T, Pesenti, E, Galvani, A, Isacchi, A, Colotta, F, Donati, D, Moll, J. | | Deposit date: | 2010-03-17 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Targeting the Mitotic Checkpoint for Cancer Therapy with Nms-P715, an Inhibitor of Mps1 Kinase.
Cancer Res., 70, 2010
|
|
2WW4
 
 | | a triclinic crystal form of E. coli 4-diphosphocytidyl-2C-methyl-D- erythritol kinase | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL | | Authors: | Kalinowska-Tluscik, J, Miallau, L, Gabrielsen, M, Leonard, G.A, McSweeney, S.M, Hunter, W.N. | | Deposit date: | 2009-10-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triclinic Crystal Form of Escherichia Coli 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase and Reassessment of the Quaternary Structure.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2WVF
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
2Z2D
 
 | |
6RT0
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
3I3V
 
 | |
2Z4E
 
 | |
2WS4
 
 | | Semi-synthetic analogue of human insulin ProB26-DTI in monomer form | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WS0
 
 | | Semi-synthetic analogue of human insulin NMeAlaB26-insulin at pH 7.5 | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WYJ
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Z6M
 
 | |
3BWL
 
 | | Crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, MAGNESIUM ION, Sensor protein | | Authors: | Osipiuk, J, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui.
To be Published
|
|
3BX7
 
 | |
2X2G
 
 | | CRYSTALLOGRAPHIC BINDING STUDIES WITH AN ENGINEERED MONOMERIC VARIANT OF TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X1U
 
 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
3C5X
 
 | | Crystal structure of the precursor membrane protein- envelope protein heterodimer from the dengue 2 virus at low pH | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]alpha-D-altropyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The flavivirus precursor membrane-envelope protein complex: structure and maturation.
Science, 319, 2008
|
|
2WVC
 
 | | Structural and mechanistic insights into Helicobacter pylori NikR function | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR, ... | | Authors: | Dian, C, Bahlawane, C, Muller, C, Round, A, Delay, C, Fauquant, C, Schauer, K, de Reuse, H, Michaud-Soret, I, Terradot, L. | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights Into Helicobacter Pylori Nikr Activation.
Nucleic Acids Res., 38, 2010
|
|
3BLW
 
 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3BOE
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 with acetate (CDCA1-R2) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BLA
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG153249 | | Descriptor: | 5-[(1S)-1-(3-chlorophenyl)ethoxy]quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
2X6S
 
 | | Human foamy virus integrase - catalytic core. Magnesium-bound structure. | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
