6Y6K
 
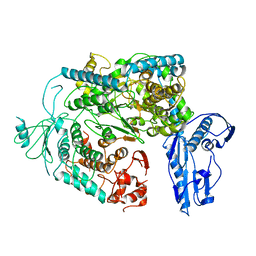 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
6Y6Z
 
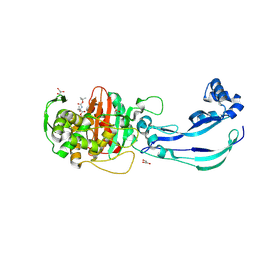 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
6Y7F
 
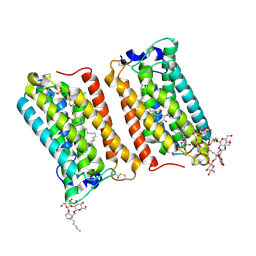 | | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7) | | Descriptor: | CHLORIDE ION, Elongation of very long chain fatty acids protein 7, Octyl Glucose Neopentyl Glycol, ... | | Authors: | Nie, L, Pike, A.C.W, Bushell, S.R, Chu, A, Cole, V, Speedman, D, Rodstrom, K.E.J, Kupinska, K, Shrestha, L, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7)
TO BE PUBLISHED
|
|
6LA8
 
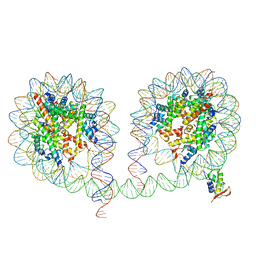 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
5CMV
 
 | | Ultrafast dynamics in myoglobin: dark-state, CO-ligated structure | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
5CND
 
 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6Y23
 
 | |
5CN5
 
 | | Ultrafast dynamics in myoglobin: 0 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6UCT
 
 | | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Klinke, S, Monti, D, Sycz, G, Cerutti, M.L, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2019-09-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant)
To Be Published
|
|
6UD0
 
 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
6UG1
 
 | | Sequence impact in DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.
DNA Repair (Amst), 107, 2021
|
|
5C14
 
 | | Crystal structure of PECAM-1 D1D2 domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Zhou, D, Paddock, C, Newman, P, Zhu, J. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for PECAM-1 homophilic binding.
Blood, 127, 2016
|
|
6YKI
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_092 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-ethyl-2-[(2~{S},5~{R})-5-methyl-2-phenyl-morpholin-4-yl]ethanamine | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
5CCN
 
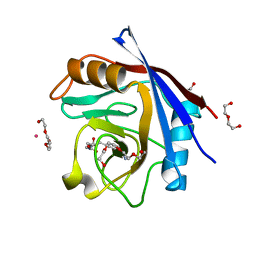 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
6YLT
 
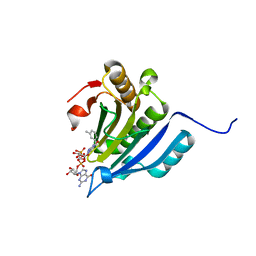 | | Translation initiation factor 4E in complex with 3-MeBn7GpppG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, [[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-methylphenyl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6LEU
 
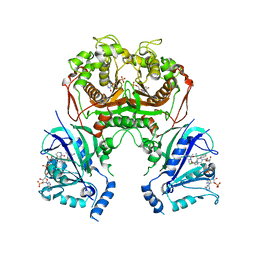 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 42 and NADPH | | Descriptor: | 1-[3-[(4-chlorophenyl)-(phenylmethyl)amino]propoxy]-6,6-dimethyl-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
6YS3
 
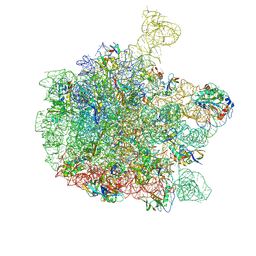 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
6TV7
 
 | | Crystal structure of rsGCaMP in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, SODIUM ION, rsGCaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-01-09 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
5CIO
 
 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
5C6R
 
 | | Crystal structure of PH domain of ASAP1 | | Descriptor: | Arf-GAP, PHOSPHATE ION, TRIETHYLENE GLYCOL | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1.
Structure, 23, 2015
|
|
6L2N
 
 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-3bp-GTAC) complex | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*GP*TP*AP*CP*TP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*GP*A)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
5C84
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 20 | | Descriptor: | (2R,5R)-4-[2-(6-chloro-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6Y5R
 
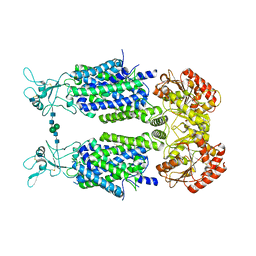 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, B, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure of Human Potassium Chloride Transporter KCC3 in NaCl
To be published
|
|
