4H0J
 
 | | Mutant M58C of Nostoc sp Cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Pannu, N.S, Skubak, P, Ubbink, M, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2012-09-08 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
2Z8D
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
3WF9
 
 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate | | Descriptor: | (2S)-1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl (2S)-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
1TRH
 
 | |
4JBS
 
 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 in complex with PHOSPHINIC PSEUDOTRIPEPTIDE inhibitor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Birtley, J, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5ZA1
 
 | | Ligand complex of RORgt LBD | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-({[4-(ethylsulfonyl)phenyl]acetyl}amino)phenyl]-2-methyl-N-phenylpropanamide, DIMETHYLFORMAMIDE, ... | | Authors: | Yamamoto, S, Yamaguchi, H. | | Deposit date: | 2018-02-06 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of a potent orally bioavailable retinoic acid receptor-related orphan receptor-gamma-t (ROR gamma t) inhibitor, S18-000003.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3PIH
 
 | | T. maritima UvrA in complex with fluorescein-modified DNA | | Descriptor: | DNA (32-MER), PYROPHOSPHATE, UvrABC system protein A, ... | | Authors: | Jaciuk, M, Nowak, E, Nowotny, M. | | Deposit date: | 2010-11-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of UvrA nucleotide excision repair protein in complex with modified DNA.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6BKL
 
 | | Influenza A M2 transmembrane domain bound to rimantadine | | Descriptor: | (1S)-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-amine, Matrix protein 2, RIMANTADINE | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters.
J. Am. Chem. Soc., 140, 2018
|
|
4IBM
 
 | | Crystal structure of insulin receptor kinase domain in complex with an inhibitor Irfin-1 | | Descriptor: | 5-(2-phenylpyrazolo[1,5-a]pyridin-3-yl)-3H-pyrazolo[3,4-c]pyridazin-3-one, Insulin receptor | | Authors: | Wu, J, Anastassiadis, T, Duong-Ly, K.C, Peterson, J.R. | | Deposit date: | 2012-12-08 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A highly selective dual insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF-1R) inhibitor derived from an extracellular signal-regulated kinase (ERK) inhibitor.
J.Biol.Chem., 288, 2013
|
|
6BKK
 
 | | Influenza A M2 transmembrane domain bound to amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters.
J. Am. Chem. Soc., 140, 2018
|
|
6E0R
 
 | | hALK in complex with compound 7 N-((1S)-1-(5-fluoropyridin-2-yl)ethyl)-1-(5-methyl-1H-pyrazol-3-yl)-3-(oxetan-3-ylsulfonyl)-1H-pyrrolo[2,3-b]pyridin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(5-fluoropyridin-2-yl)ethyl]-1-(5-methyl-1H-pyrazol-3-yl)-3-[(oxetan-3-yl)sulfonyl]-1H-pyrrolo[2,3-b]pyridin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-07-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6EDL
 
 | | hALK in complex with compound 1 (S)-N-(1-(2,4-difluorophenyl)ethyl)-3-(3-methyl-1H-pyrazol-5-yl)imidazo[1,2-b]pyridazin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(2,4-difluorophenyl)ethyl]-3-(5-methyl-1H-pyrazol-3-yl)imidazo[1,2-b]pyridazin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
1FMC
 
 | | 7-ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEX WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1996-04-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
6EBW
 
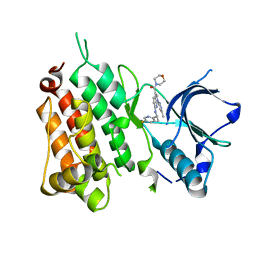 | | hALK in complex with compound 9 (6-(((1S)-1-(5-Fluoropyridin-2-yl)ethyl)amino)-1-(3-methyl-1H-pyrazol-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl)(morpholin-4-yl)methanone | | Descriptor: | ALK tyrosine kinase receptor, [6-{[(1S)-1-(5-fluoropyridin-2-yl)ethyl]amino}-1-(5-methyl-1H-pyrazol-3-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl](morpholin-4-yl)methanone | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-08-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6BUV
 
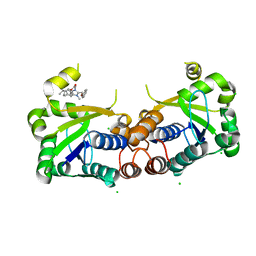 | | Structure of Mycobacterium tuberculosis NadD in complex with inhibitor [(1~{R},2~{R},5~{S})-5-methyl-2-propan-2-yl-cyclohexyl] 2-[3-methyl-2-(phenoxymethyl)benzimidazol-1-yl]ethanoate | | Descriptor: | 1-methyl-3-(2-{[(1R,2R,5S)-5-methyl-2-(propan-2-yl)cyclohexyl]oxy}-2-oxoethyl)-2-(phenoxymethyl)-1H-1,3-benzimidazol-3-ium, CHLORIDE ION, SODIUM ION, ... | | Authors: | Rodionova, I.A, Reed, R.W, Sorci, L, Osterman, A.L, Korotkov, K.V. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Novel Antimycobacterial Compounds Suppress NAD Biogenesis by Targeting a Unique Pocket of NaMN Adenylyltransferase.
Acs Chem.Biol., 14, 2019
|
|
4CRL
 
 | |
1ND3
 
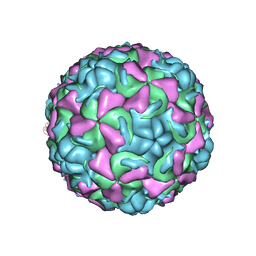 | | The structure of HRV16, when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, ZINC ION, coat protein VP1, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
4YPD
 
 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
1NCR
 
 | | The structure of Rhinovirus 16 when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, MYRISTIC ACID, ZINC ION, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-05 | | Release date: | 2003-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
1ND2
 
 | | The structure of Rhinovirus 16 | | Descriptor: | MYRISTIC ACID, ZINC ION, coat protein VP1, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
1R9H
 
 | |
4F9Y
 
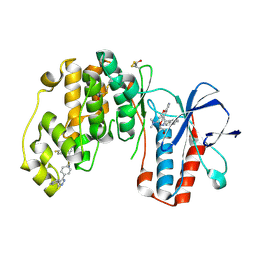 | | Human P38 alpha MAPK In Complex With a Novel and Selective Small Molecule Inhibitor | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2012-05-21 | | Release date: | 2013-06-05 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Novel In Vivo Chemical Probes to Address CNS Protein Kinase Involvement in Synaptic Dysfunction.
Plos One, 8, 2013
|
|
7C3N
 
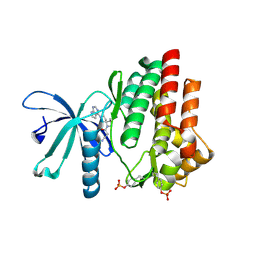 | | Crystal structure of JAK3 in complex with Delgocitinib | | Descriptor: | 3-[(3S,4R)-3-methyl-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,7-diazaspiro[3.4]octan-1-yl]-3-oxidanylidene-propanenitrile, Tyrosine-protein kinase JAK3 | | Authors: | Doi, S, Otira, T, Kikuwaka, M, Nomura, A, Noji, S, Adachi, T. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders.
J.Med.Chem., 63, 2020
|
|
7LYV
 
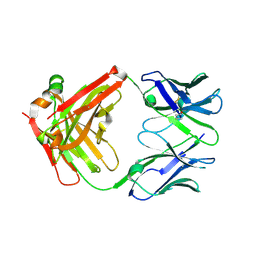 | |
7LYW
 
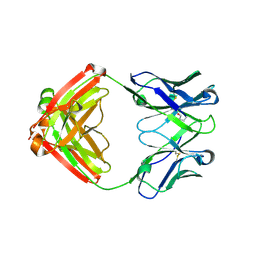 | |
