4CO4
 
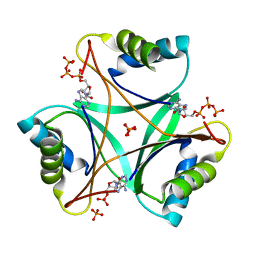 | | Structure of PII signaling protein GlnZ from Azospirillum brasilense in complex with adenosine triphosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Truan, D, Li, X.-D, Winkler, F.K. | | Deposit date: | 2014-01-25 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Thermodynamics of Effector Molecule Binding to the Nitrogen Signal Transduction Pii Protein Glnz from Azospirillum Brasilense.
J.Mol.Biol., 426, 2014
|
|
4CO0
 
 | |
4CNZ
 
 | |
4CO1
 
 | |
4CNY
 
 | |
4CO2
 
 | |
4LBB
 
 | |
4LB7
 
 | |
2ICJ
 
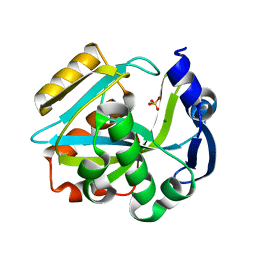 | | The crystal structure of human isopentenyl diphophate isomerase | | Descriptor: | Isopentenyl-diphosphate delta isomerase, MAGNESIUM ION, SULFATE ION | | Authors: | Zheng, W, Bartlam, M, Rao, Z. | | Deposit date: | 2006-09-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of human isopentenyl diphosphate isomerase at 1.7 A resolution reveals its catalytic mechanism in isoprenoid biosynthesis
J.Mol.Biol., 366, 2007
|
|
5XVF
 
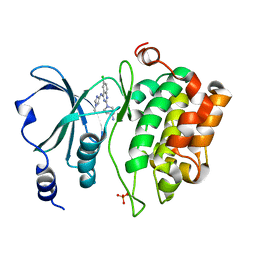 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | Descriptor: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5X8G
 
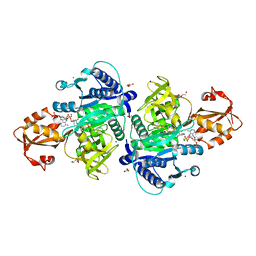 | |
6Y86
 
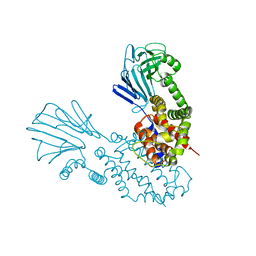 | |
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
7FAT
 
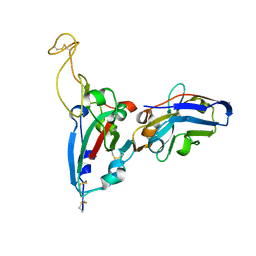 | | Structure Determination of the RBD-NB1A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | Authors: | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
7FCP
 
 | |
7FCQ
 
 | |
8ZWF
 
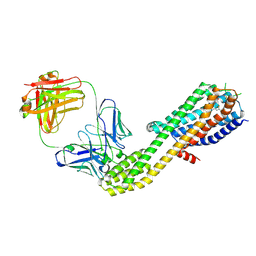 | | cryoEM structure of JR14a bound C3aR-BRIL-BAG2 complex | | Descriptor: | (2~{S})-5-[bis(azanyl)methylideneamino]-2-[[5-[bis(4-chlorophenyl)methyl]-3-methyl-thiophen-2-yl]carbonylamino]pentanoic acid, Beta-2 adrenergic receptor,C3a anaphylatoxin chemotactic receptor,Soluble cytochrome b562, anti-BRIL Fab HC, ... | | Authors: | Luo, P, Xu, Y, Xu, H.E. | | Deposit date: | 2024-06-13 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the agonist activity of the nonpeptide modulator JR14a on C3aR.
Cell Discov, 11, 2025
|
|
8ZWG
 
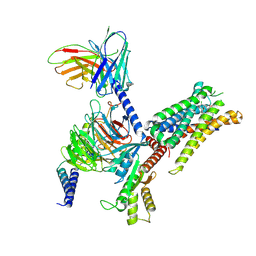 | | cryoEM structure of JR14a bound C3aR-Gi complex | | Descriptor: | (2~{S})-5-[bis(azanyl)methylideneamino]-2-[[5-[bis(4-chlorophenyl)methyl]-3-methyl-thiophen-2-yl]carbonylamino]pentanoic acid, Beta-2 adrenergic receptor,C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Luo, P, Xu, Y, Xu, H.E. | | Deposit date: | 2024-06-13 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the agonist activity of the nonpeptide modulator JR14a on C3aR.
Cell Discov, 11, 2025
|
|
4U97
 
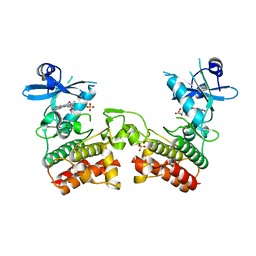 | | Crystal Structure of Asymmetric IRAK4 Dimer | | Descriptor: | Interleukin-1 receptor-associated kinase 4, STAUROSPORINE, SULFATE ION | | Authors: | Ferrao, R, Wu, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | IRAK4 Dimerization and trans-Autophosphorylation Are Induced by Myddosome Assembly.
Mol.Cell, 55, 2014
|
|
4U9A
 
 | |
3DOY
 
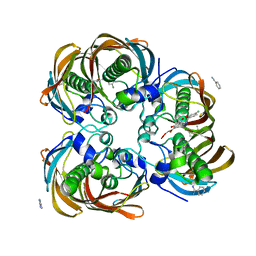 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3i | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-chloro-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP3
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3q | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-tert-butyl-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP2
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3j | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-bromo-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DOZ
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3k | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3-bromo-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP1
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3n | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
