4J46
 
 | | Crystal structure of XIAP-BIR2 domain with AVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J44
 
 | | Crystal structure of XIAP-BIR2 domain with AIAV bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-ILE-ALA-VAL), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
4J47
 
 | | Crystal structure of XIAP-BIR2 domain with SVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (SER-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J3Y
 
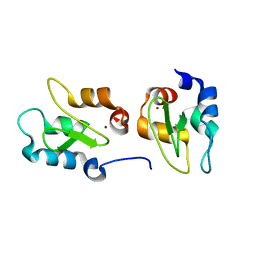 | | Crystal structure of XIAP-BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ESF
 
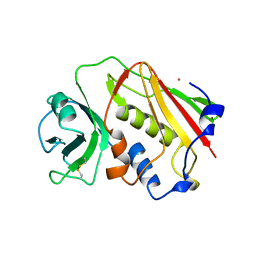 | | STAPHYLOCOCCAL ENTEROTOXIN A | | Descriptor: | CADMIUM ION, STAPHYLOCOCCAL ENTEROTOXIN A | | Authors: | Schad, E.M, Svensson, L.A. | | Deposit date: | 1995-05-25 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superantigen staphylococcal enterotoxin type A.
EMBO J., 14, 1995
|
|
4HZD
 
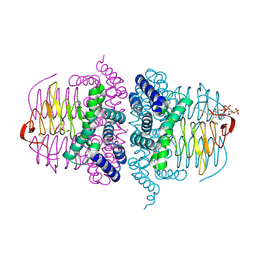 | |
4HZC
 
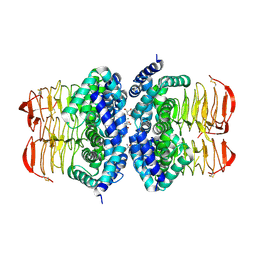 | |
4O97
 
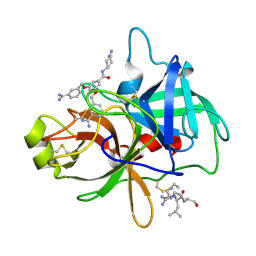 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4O9V
 
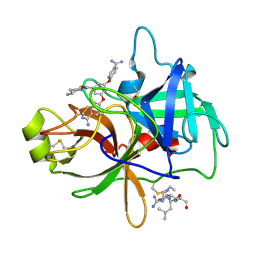 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis(4-carbamimidoylphenoxy)benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4R0I
 
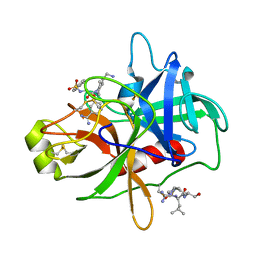 | | CRYSTAL STRUCTURE of MATRIPTASE in COMPLEX WITH INHIBITOR | | Descriptor: | 3-({(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-[(naphthalen-2-ylsulfonyl)amino]-3-oxopropyl}oxy)benzenecarboximidamide, SERINE PROTEASE, MATRIPTASE, ... | | Authors: | Rao, K.N, Ashok, K.N, Chakshusmathi, G, Rajeev, G, Subramanya, H. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1F9I
 
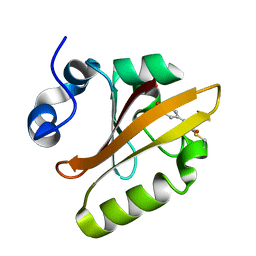 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT Y42F | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-10 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
7N1Z
 
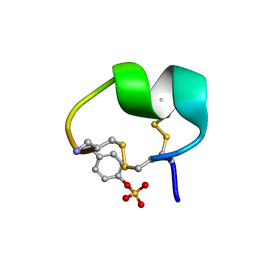 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N24
 
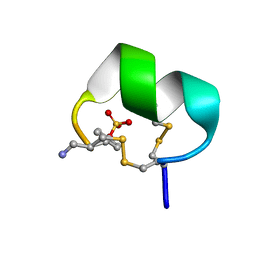 | | NMR structure of native EpI | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N25
 
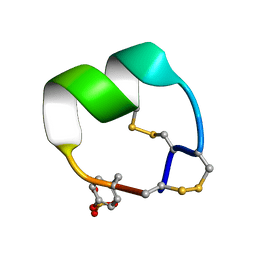 | | NMR structure of EpI-OH | | Descriptor: | Alpha-conotoxin EpI-OH | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N26
 
 | | NMR structure of EpI-[Y(SO3)15Y]-NH2 | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
8T2J
 
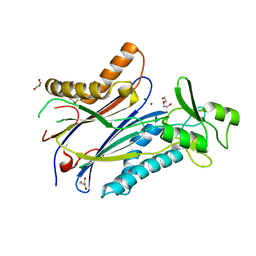 | | Structure of the catalytic domain of PPM1D/Wip1 serine/threonine phosphatase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumar, J.P, Kosek, D, Dyda, F. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanistic studies of the PPM1D serine/threonine phosphatase catalytic domain.
J.Biol.Chem., 300, 2024
|
|
1F98
 
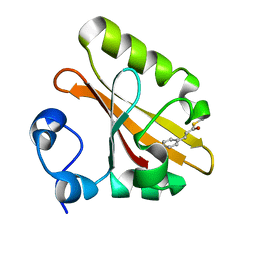 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT T50V | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-07 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
7N21
 
 | | NMR structure of AnIB-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N23
 
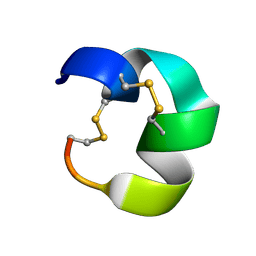 | | NMR structure of AnIB[Y(SO3)16Y]-OH | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N22
 
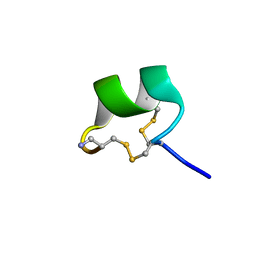 | | NMR structure of AnIB[Y(SO3)16Y]-NH2 | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N0T
 
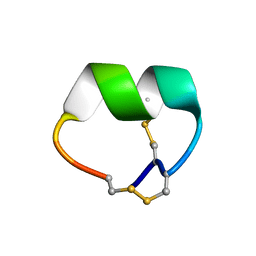 | | NMR structure of EpI[Y(SO)315Y]-OH | | Descriptor: | Alpha-conotoxin EpI | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7N20
 
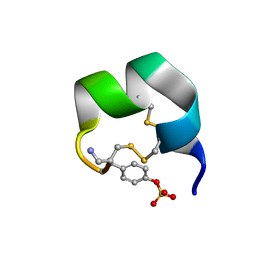 | | NMR structure of native AnIB | | Descriptor: | Alpha-conotoxin AnIB | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
6B09
 
 | |
4WTD
 
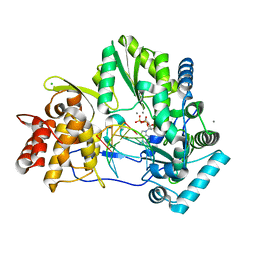 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH ADP, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-AUAAAUUU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C. | | Deposit date: | 2014-10-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
