8HR7
 
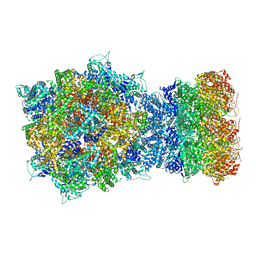 | | Structure of RdrA-RdrB complex | | Descriptor: | Adenosine deaminase, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRC
 
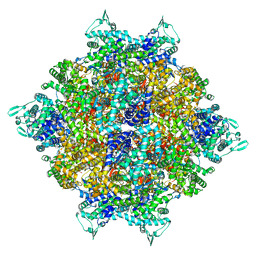 | | Structure of dodecameric RdrB cage | | Descriptor: | Adenosine deaminase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR9
 
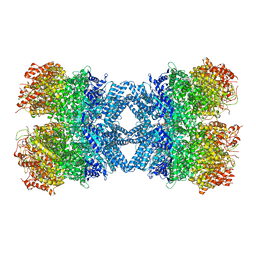 | | Structure of tetradecameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR8
 
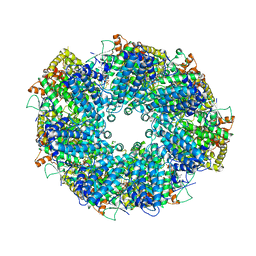 | | Structure of heptameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRA
 
 | | Structure of heptameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(P*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
5Z3R
 
 | | Crystal Structure of Delta 5-3-Ketosteroid Isomerase from Mycobacterium sp. | | Descriptor: | Steroid delta-isomerase | | Authors: | Cheng, X.Y, Peng, F, Yang, F, Huang, Y.Q, Su, Z.D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-based reconstruction of a Mycobacterium hypothetical protein into an active Delta5-3-ketosteroid isomerase.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
8HKQ
 
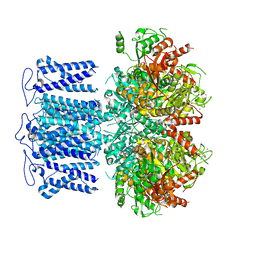 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKK
 
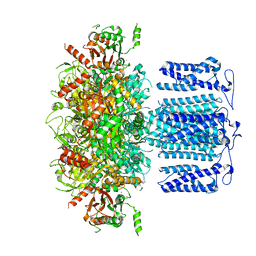 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HIR
 
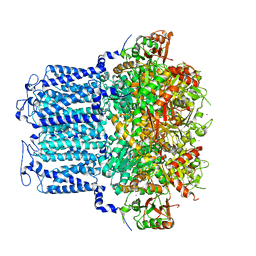 | | potassium channels | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKM
 
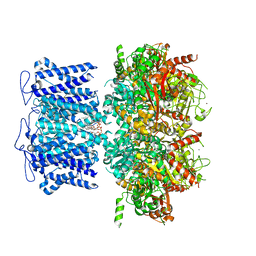 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HK6
 
 | | potassium channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKF
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, Z.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
6DMJ
 
 | | A multiconformer ligand model of inhibitor 53W bound to CREB binding protein bromodomain | | Descriptor: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[2-(4-methoxyphenyl)ethyl]-1-[2-(morpholin-4-yl)ethyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
5XHS
 
 | | Crystal structure of SIRT5 complexed with a fluorogenic small-molecule substrate SuBKA | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, 7-AMINO-4-METHYL-CHROMEN-2-ONE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Yu, Y, Li, B, Chen, Q. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions between sirtuins and fluorogenic small-molecule substrates offer insights into inhibitor design
Rsc Adv, 7, 2017
|
|
6KYB
 
 | | Crystal structure of Atg18 from Saccharomyces cerevisiae | | Descriptor: | Autophagy-related protein 18 | | Authors: | Tang, D, Lei, Y, Liao, G, Chen, Q, Xu, L, Lu, K, Qi, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of Atg18 reveals a new binding site for Atg2 in Saccharomyces cerevisiae.
Cell.Mol.Life Sci., 78, 2021
|
|
6IIC
 
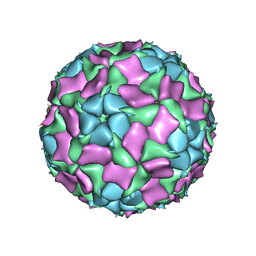 | | CryoEM structure of Mud Crab Dicistrovirus | | Descriptor: | VP1 of Mud crab dicistrovirus, VP2 of Mud crab dicistrovirus, VP3 of Mud crab dicistrovirus, ... | | Authors: | Zhang, Q, Gao, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Novel Viruses from Mud CrabScylla paramamosainwith Multiple Infections.
J. Virol., 93, 2019
|
|
6JJI
 
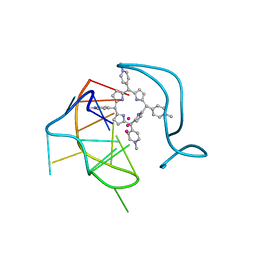 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 (1:1) | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJH
 
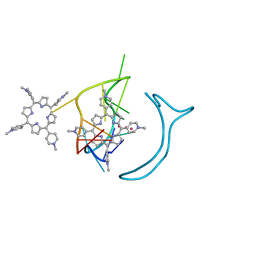 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JO8
 
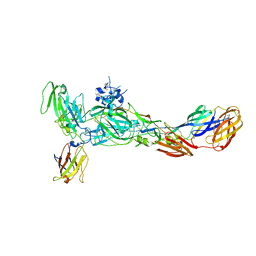 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JJF
 
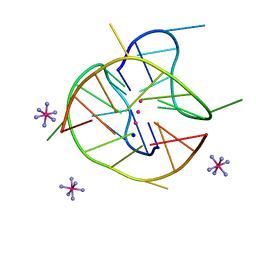 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6IZL
 
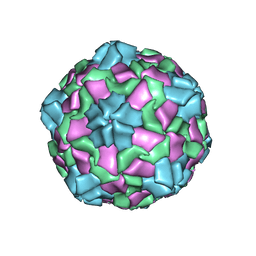 | |
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
7E0M
 
 | | Crystal structure of phospholipase D | | Descriptor: | Phospholipase, SULFATE ION | | Authors: | Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a Phospholipase D from the Plant-Associated Bacteria Serratia plymuthica Strain AS9 Reveals a Unique Arrangement of Catalytic Pocket.
Int J Mol Sci, 22, 2021
|
|
7F8G
 
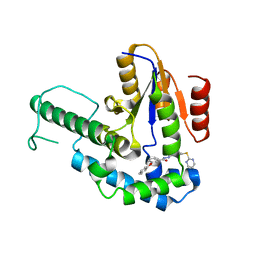 | |
