4D81
 
 | | Metallosphera sedula Vps4 crystal structure | | Descriptor: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
4DSO
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DST
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2R17
 
 | | Functional architecture of the retromer cargo-recognition complex | | Descriptor: | GLYCEROL, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Hierro, A, Rojas, A.L, Rojas, R, Murthy, N, Effantin, G, Kajava, A.V, Steven, A.C, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2007-08-22 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional architecture of the retromer cargo-recognition complex.
Nature, 449, 2007
|
|
7ZCH
 
 | | CHMP2A-CHMP3 heterodimer (410 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZCG
 
 | | CHMP2A-CHMP3 heterodimer (430 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5T84
 
 | | HIV-1 protease, unbound subtype B L63P construct | | Descriptor: | Protease | | Authors: | Liu, Z, Norell, J, Mahon, B, Poole, K, McKenna, R, Fanucci, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single mutation, L63P, capable of shutting down the flaps of HIV-1 protease
To Be Published
|
|
7K7G
 
 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K79
 
 | | CBF3 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K78
 
 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
5Z6Q
 
 | | Crystal structure of AAA of Spastin | | Descriptor: | CHLORIDE ION, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The AAA protein spastin possesses two levels of basal ATPase activity
FEBS Lett., 592, 2018
|
|
5NC5
 
 | |
5O66
 
 | | Asymmetric AcrABZ-TolC | | Descriptor: | Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump.
Elife, 6, 2017
|
|
8XEP
 
 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | Descriptor: | SULFATE ION, Type IV effector MavL, Ubiquitin | | Authors: | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
7EZT
 
 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-06-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
8EC0
 
 | | III2IV respiratory supercomplex from Saccharomyces cerevisiae cardiolipin-lacking mutant | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Hryc, C.F, Mileykovskaya, E, Baker, M, Dowhan, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into cardiolipin replacement by phosphatidylglycerol in a cardiolipin-lacking yeast respiratory supercomplex.
Nat Commun, 14, 2023
|
|
8E7S
 
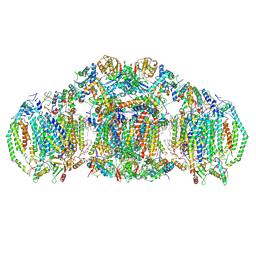 | | III2IV2 respiratory supercomplex from Saccharomyces cerevisiae with 4 bound UQ6 | | Descriptor: | (5S,11R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-4,6,10,12,16-pentaoxa-5,11-diphosphaoctadec-1-yl pentadecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Hryc, C.F, Mileykovskaya, E, Baker, M, Dowhan, W. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into cardiolipin replacement by phosphatidylglycerol in a cardiolipin-lacking yeast respiratory supercomplex.
Nat Commun, 14, 2023
|
|
6BO4
 
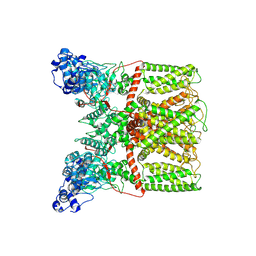 | |
6BO5
 
 | | TRPV2 ion channel in partially closed state | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Dosey, T.L, Wang, Z. | | Deposit date: | 2017-11-18 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of TRPV2 in distinct conformations provide insight into role of the pore turret.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8JO4
 
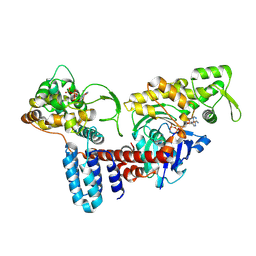 | | Cryo-EM structure of a Legionella effector complexed with actin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8JO3
 
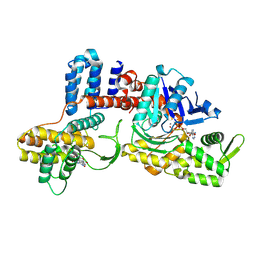 | | Cryo-EM structure of a Legionella effector complexed with actin and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
3GOL
 
 | | HCV NS5b polymerase in complex with 1,5 benzodiazepine inhibitor (R)-11d | | Descriptor: | (11R)-10-acetyl-11-(2,4-dichlorophenyl)-6-hydroxy-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Nyanguile, O, De Bondt, H. | | Deposit date: | 2009-03-19 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 1,5-Benzodiazepine inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4DRU
 
 | | HCV NS5B in complex with macrocyclic INDOLE INHIBITOR | | Descriptor: | 13-cyclohexyl-3-methoxy-17,22-dimethyl-7H-10,6-(methanoiminothioiminobutanoiminomethano)indolo[2,1-a][2]benzazepine-14,23-dione 16,16-dioxide, GLYCEROL, RNA-directed RNA polymerase, ... | | Authors: | Cummings, M.D, Vendeville, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based macrocyclization yields hepatitis C virus NS5B inhibitors with improved binding affinities and pharmacokinetic properties.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
