1G6T
 
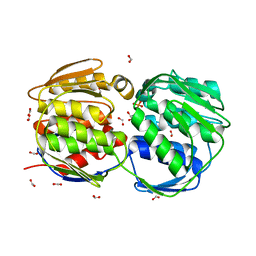 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G6S
 
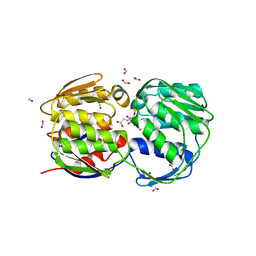 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE AND GLYPHOSATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, GLYPHOSATE, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7WU5
 
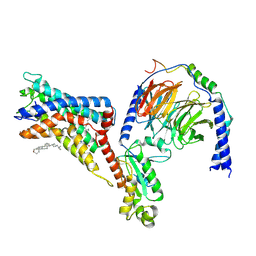 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
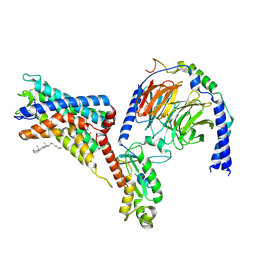 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU3
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU2
 
 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | Authors: | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | Deposit date: | 2017-10-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
8Z4F
 
 | |
7DAE
 
 | | EPB in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, CALCIUM ION, ... | | Authors: | Wu, C, Wang, Y. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7DAF
 
 | | IXA in complex with tubulin | | Descriptor: | (1~{S},3~{S},7~{S},10~{R},11~{S},12~{S},16~{R})-8,8,10,12,16-pentamethyl-3-[(~{E})-1-(2-methyl-1,3-thiazol-4-yl)prop-1-en-2-yl]-7,11-bis(oxidanyl)-17-oxa-4-azabicyclo[14.1.0]heptadecane-5,9-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wu, C, Wang, Y. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7DAD
 
 | | EPD in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y, Wu, C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
4JEK
 
 | | Structure of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | Dibenzothiophene desulfurization enzyme C | | Authors: | Zhang, L, Duan, X.L, Zhou, D.M, Li, X. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary structural analysis of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3S0Z
 
 | | Crystal structure of New Delhi Metallo-beta-lactamase (NDM-1) | | Descriptor: | Metallo-beta-lactamase, ZINC ION | | Authors: | Guo, Y, Wang, J, Niu, G.J, Shui, W.Q, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural view of the antibiotic degradation enzyme NDM-1 from a superbug.
Protein Cell, 2011
|
|
3UIM
 
 | |
