6I6H
 
 | |
6I6J
 
 | |
7B0K
 
 | | membrane protein structure | | Descriptor: | CHOLINE ION, Drug/metabolite transporter (DMT) superfamily permease | | Authors: | Baerland, N, Perez, C. | | Deposit date: | 2020-11-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification.
Sci Adv, 8, 2022
|
|
7A29
 
 | |
7PAF
 
 | | Streptococcus pneumoniae choline importer LicB in lipid nanodiscs | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, LicB protein, Nanobody | | Authors: | Perez, C, Baerland, N. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification.
Sci Adv, 8, 2022
|
|
6QGX
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGW
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody E6 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoE6, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
7NDF
 
 | |
4U8Y
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB.
Elife, 3, 2014
|
|
4U95
 
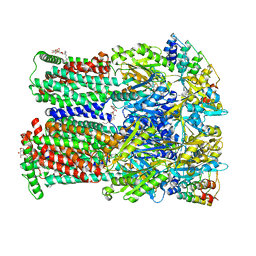 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB
eLife, 3, 2014
|
|
4U96
 
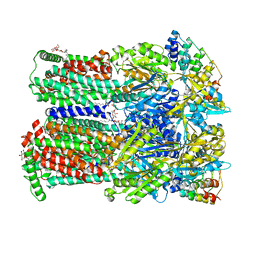 | |
4U8V
 
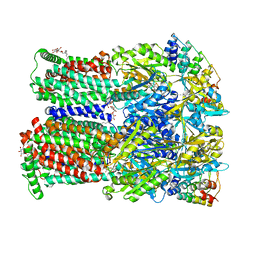 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB.
Elife, 3, 2014
|
|
3NOG
 
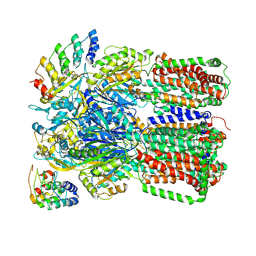 | |
3NOC
 
 | |
8OMZ
 
 | |
8OO1
 
 | |
8PD0
 
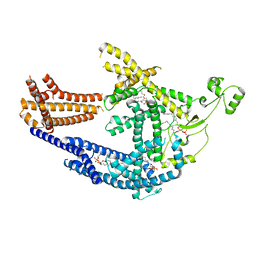 | | cryo-EM structure of Doa10 in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PDA
 
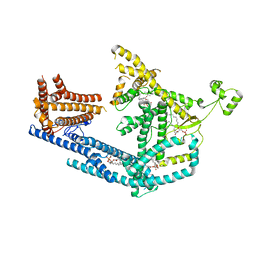 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7P60
 
 | |
7NGH
 
 | | Structure of glutamate transporter homologue in complex with Sybody | | Descriptor: | ASPARTIC ACID, Proton/glutamate symporter, SDF family, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Kinetic mechanism of Na + -coupled aspartate transport catalyzed by Glt Tk .
Commun Biol, 4, 2021
|
|
7P5W
 
 | |
