3H9U
 
 | | S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Siponen, M.I, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh Nielsen, T, Kotenyova, T, Kotzsch, A, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei
To be Published
|
|
1MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
6TBM
 
 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | Descriptor: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-11-01 | | Release date: | 2020-02-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6TB4
 
 | | Structure of SAGA bound to TBP | | Descriptor: | SAGA-associated factor 73 (Sgf73), Spt20, Subunit (17 kDa) of TFIID and SAGA complexes, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
230D
 
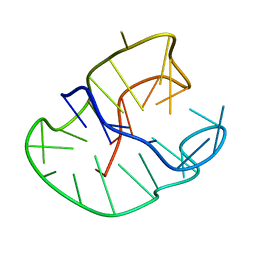 | |
2AVW
 
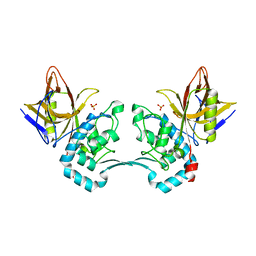 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | Descriptor: | GLYCEROL, IgG-degrading protease, SULFATE ION | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
6HQA
 
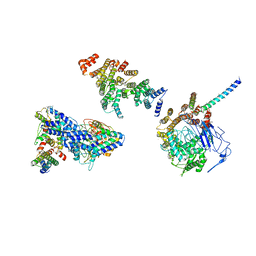 | | Molecular structure of promoter-bound yeast TFIID | | Descriptor: | Histone-fold, Subunit (17 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation, ... | | Authors: | Kolesnikova, O, Ben-Shem, A, Luo, J, Ranish, J, Schultz, P, Papai, G. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular structure of promoter-bound yeast TFIID.
Nat Commun, 9, 2018
|
|
2AU1
 
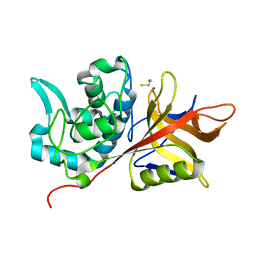 | | Crystal Structure of group A Streptococcus MAC-1 orthorhombic form | | Descriptor: | BETA-MERCAPTOETHANOL, IgG-degrading protease | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-26 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2QMI
 
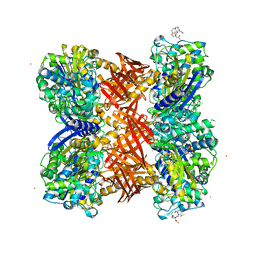 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
1D3X
 
 | | INTRAMOLECULAR DNA TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Tarkoy, M, Phipps, A.K, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex linked by hexakis(ethylene glycol) units: d(AGAGAGAA-(EG)6-TTCTCTCT-(EG)6-TCTCTCTT).
Biochemistry, 37, 1998
|
|
1CFF
 
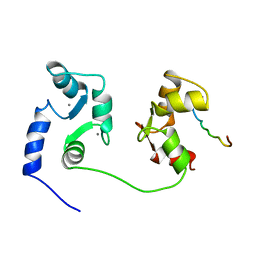 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
1DJ7
 
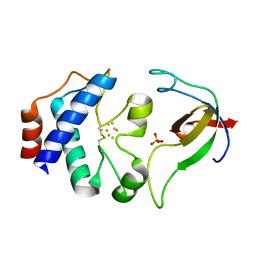 | | CRYSTAL STRUCTURE OF FERREDOXIN THIOREDOXIN REDUCTASE | | Descriptor: | FERREDOXIN THIOREDOXIN REDUCTASE: CATALYTIC CHAIN, FERREDOXIN THIOREDOXIN REDUCTASE: VARIABLE CHAIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dai, S, Schwendtmayer, C, Schurmann, P, Ramaswamy, S, Eklund, H. | | Deposit date: | 1999-12-02 | | Release date: | 2000-02-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox signaling in chloroplasts: cleavage of disulfides by an iron-sulfur cluster.
Science, 287, 2000
|
|
2PUK
 
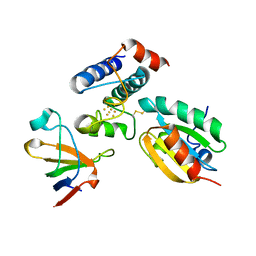 | | Crystal structure of the binary complex between ferredoxin: thioredoxin reductase and thioredoxin m | | Descriptor: | Ferredoxin-thioredoxin reductase, catalytic chain, variable chain, ... | | Authors: | Dai, S, Friemann, R, Schurmann, P, Eklund, H. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural snapshots along the reaction pathway of ferredoxin-thioredoxin reductase.
Nature, 448, 2007
|
|
1R3X
 
 | | INTRAMOLECULAR DNA TRIPLEX WITH RNA THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3'), RNA (5'-R(*UP*CP*UP*CP*UP*CP*UP*U)-3') | | Authors: | Gotfredsen, C.H, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Intramolecular Pyrimidine-Purine-Pyrimidine Triplex Containing an RNA Third Strand
J.Am.Chem.Soc., 120, 1998
|
|
1CG7
 
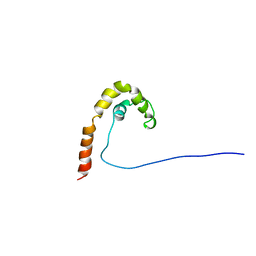 | | HMG PROTEIN NHP6A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PROTEIN (NON HISTONE PROTEIN 6 A) | | Authors: | Allain, F.H.T, Yen, Y.M, Masse, J.E, Schultze, P, Dieckmann, T, Johnson, R.C, Feigon, J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding.
EMBO J., 18, 1999
|
|
1P3X
 
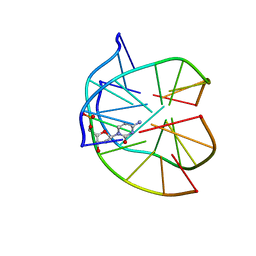 | | INTRAMOLECULAR DNA TRIPLEX WITH 1-PROPYNYL DEOXYURIDINE IN THE THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*(PDU)P*CP*(PDU)P*(DCM)P*(PDU)P*CP*(PDU)P*(PDU))-3'), DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Phipps, A.K, Tarkoy, M, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2'-deoxyuridine residues in the third strand.
Biochemistry, 37, 1998
|
|
1SK3
 
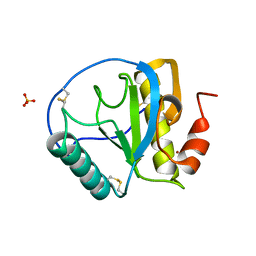 | | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | NICKEL (II) ION, Peptidoglycan recognition protein I-alpha, SULFATE ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1SK4
 
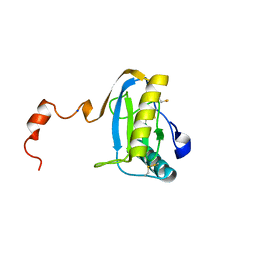 | | crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | Peptidoglycan recognition protein I-alpha, SODIUM ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1EFX
 
 | | STRUCTURE OF A COMPLEX BETWEEN THE HUMAN NATURAL KILLER CELL RECEPTOR KIR2DL2 AND A CLASS I MHC LIGAND HLA-CW3 | | Descriptor: | BETA-2-MICROGLOBULIN, HLA-CW3 (HEAVY CHAIN), NATURAL KILLER CELL RECEPTOR KIR2DL2, ... | | Authors: | Boyington, J.C, Motyka, S.A, Schuck, P, Brooks, A.G, Sun, P.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand.
Nature, 405, 2000
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
5OEJ
 
 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | Descriptor: | Tra1 subunit within the chromatin modifying complex SAGA | | Authors: | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
1F9M
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (SHORT FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-11 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FB0
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN M FROM SPINACH CHLOROPLAST (REDUCED FORM) | | Descriptor: | THIOREDOXIN M | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-14 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FB6
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN M FROM SPINACH CHLOROPLAST (OXIDIZED FORM) | | Descriptor: | THIOREDOXIN M | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-14 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
1FAA
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN F FROM SPINACH CHLOROPLAST (LONG FORM) | | Descriptor: | THIOREDOXIN F | | Authors: | Capitani, G, Markovic-Housley, Z, DelVal, G, Morris, M, Jansonius, J.N, Schurmann, P. | | Deposit date: | 2000-07-13 | | Release date: | 2000-09-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two functionally different thioredoxins in spinach chloroplasts.
J.Mol.Biol., 302, 2000
|
|
