5AC1
 
 | | Sheep aldehyde dehydrogenase 1A1 with duocarmycin analog inhibitor | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, MAGNESIUM ION, RETINAL DEHYDROGENASE 1, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AC0
 
 | | ovis aries Aldehyde Dehydrogenase 1A1 in complex with a duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A3D
 
 | | Structural insights into the recognition of cisplatin and AAF-dG lesions by Rad14 (XPA) | | Descriptor: | 5'-D(*DG 5IUP*GP*A 5IUP*GP*AP*CP*G 5IUP*AP*GP*AP*DGP*AP)-3', 5'-D(*DTP*CP*TP*CP*TP*AP*C 8FGP*TP*CP*AP*TP*CP*DAP*CP)-3', DNA REPAIR PROTEIN RAD14, ... | | Authors: | Kuper, J, Koch, S.C, Gasteiger, K.L, Wichlein, N, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6G56
 
 | |
6G59
 
 | | Structure of the alanine racemase from Staphylococcus aureus in complex with an pyridoxal-6- phosphate derivative | | Descriptor: | (6-ethynyl-4-methanoyl-5-oxidanyl-pyridin-3-yl)methyl dihydrogen phosphate, Alanine racemase 1, CHLORIDE ION, ... | | Authors: | Hoegl, A, Sieber, S.A, Schneider, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mining the cellular inventory of pyridoxal phosphate-dependent enzymes with functionalized cofactor mimics.
Nat Chem, 10, 2018
|
|
2WTF
 
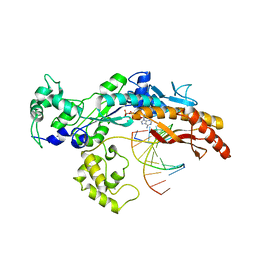 | | DNA polymerase eta in complex with the cis-diammineplatinum (II) 1,3- GTG intrastrand cross-link | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*TP*GP*GP*TP*GP*AP*GP*CP)-3', 5'-D(*TP*CP*TP*TP*CP*TP*GP*TP*GP*CP *TP*CP*AP*CP*CP*AP*CP)-3', ... | | Authors: | Reissner, T, Schneider, S, Ziv, O, Schorr, S, Livneh, Z, Carell, T. | | Deposit date: | 2009-09-16 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Cisplatin-(1,3-Gtg) Cross-Link within DNA Polymerase Eta.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6G58
 
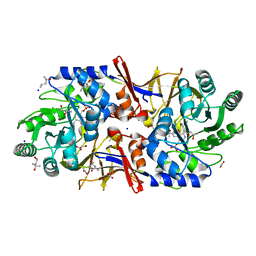 | | Structure of the alanine racemase from Staphylococcus aureus in complex with a pyridoxal 5' phosphate-derivative | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, (6-but-3-ynyl-4-methyl-5-oxidanyl-pyridin-3-yl)methyl dihydrogen phosphate, ... | | Authors: | Hoegl, A, Sieber, S.A, Schneider, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mining the cellular inventory of pyridoxal phosphate-dependent enzymes with functionalized cofactor mimics.
Nat Chem, 10, 2018
|
|
3CVV
 
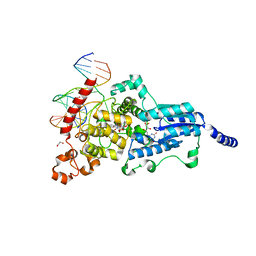 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion and F0 cofactor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), ... | | Authors: | Glas, A.F, Maul, M.J, Cryle, M.J, Barends, T.R.M, Schneider, S, Kaya, E, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WQ6
 
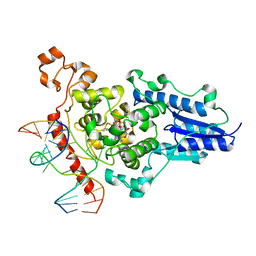 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(Dewar)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*CDWP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions
J.Am.Chem.Soc., 132, 2010
|
|
5OL1
 
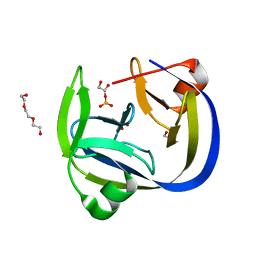 | | Crystal structure of an inactivated Ssp SICLOPPS intein with a CAFHPQ extein | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DNA polymerase III subunit alpha,DNA polymerase III subunit alpha, ... | | Authors: | Kick, L.M, Schneider, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Mechanistic Insights into Cyclic Peptide Generation by DnaE Split-Inteins through Quantitative and Structural Investigation.
Chembiochem, 18, 2017
|
|
2Y1I
 
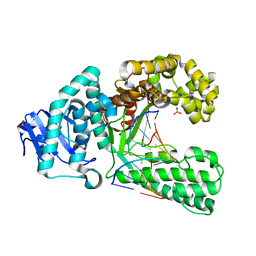 | | Crystal structure of a S-diastereomer analogue of the spore photoproduct in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*AP*GP*GP*GP*PBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
2XY5
 
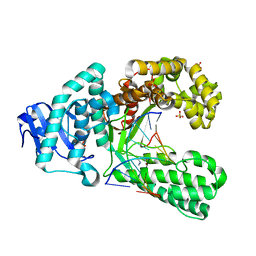 | | Crystal structure of an artificial salen-copper basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*SAYP*TP*CP*CP*CP*TP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2XY7
 
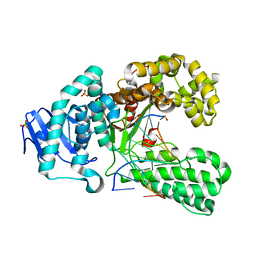 | | Crystal structure of a salicylic aldehyde base in the pre-insertion site of fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP)-3', 5'-D(*SAYP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP)-3', DNA POLYMERASE I, ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2XY6
 
 | | Crystal structure of a salicylic aldehyde basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2Y1J
 
 | | CRYSTAL STRUCTURE OF A R-DIASTEREOMER ANALOGUE OF THE SPORE PHOTOPRODUCT IN COMPLEX WITH FRAGMENT DNA POLYMERASE I FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | 5'-D(*AP*GP*GP*GP*QBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
5OL6
 
 | |
5OL5
 
 | | Crystal structure of an inactivated Ssp SICLOPPS intein with CFAHPQ extein | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DNA polymerase III subunit alpha,DNA polymerase III subunit alpha, ... | | Authors: | Kick, L.M, Schneider, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Mechanistic Insights into Cyclic Peptide Generation by DnaE Split-Inteins through Quantitative and Structural Investigation.
Chembiochem, 18, 2017
|
|
5OL7
 
 | |
2WQ7
 
 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(6-4)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*ZP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions.
J.Am.Chem.Soc., 132, 2010
|
|
2O6P
 
 | | Crystal Structure of the heme-IsdC complex | | Descriptor: | CHLORIDE ION, Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sharp, K.H, Schneider, S, Cockayne, A, Paoli, M. | | Deposit date: | 2006-12-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus.
J. Biol. Chem., 282, 2007
|
|
4B9S
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion outside of the pre-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9M
 
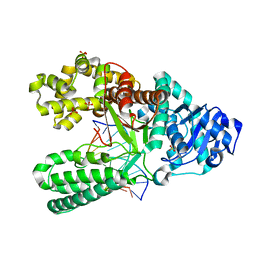 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dA DNA lesion -thymine basepair in the post- insertion site. | | Descriptor: | 5'-D(*DC*DA*DA*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9U
 
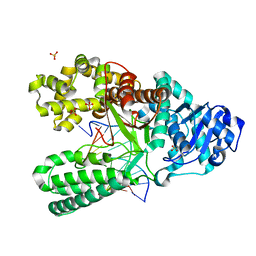 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dA basepair in the post-insertion site. | | Descriptor: | 5'-D(*CP*AP*AP*FOXP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9T
 
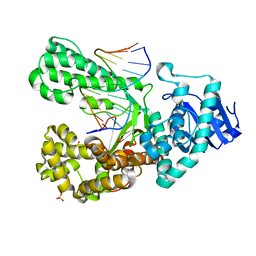 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dC basepair in the post-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
