4Q6O
 
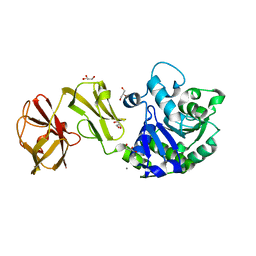 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6P
 
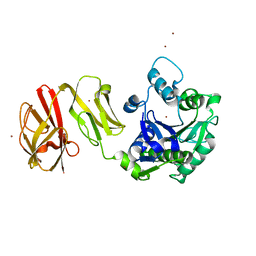 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7XHY
 
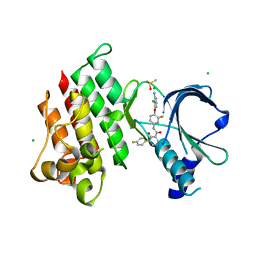 | | Crystal structure of MerTK Kinase domain with BMS794833 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Tyrosine-protein kinase Mer, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-04-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | BMS794833 inhibits macrophage efferocytosis by directly binding to MERTK and inhibiting its activity.
Exp.Mol.Med., 54, 2022
|
|
7YA9
 
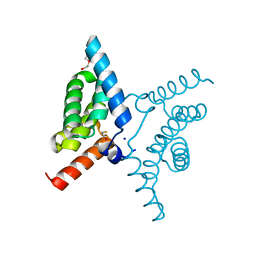 | | Fis1 (Mitochondrial fission 1 protein) | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, Mitochondrial fission 1 protein, ... | | Authors: | Duc, N.M, Bong, S.M, Lee, B.I. | | Deposit date: | 2022-06-27 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight into the Mitochondria-ER tethering revealed by the crystal structure of Fis1-Bap31 complex
To Be Published
|
|
7YKA
 
 | |
1UAL
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAJ
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAM
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAK
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
3LY6
 
 | |
1FA2
 
 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Lee, B.I, Cheong, C.G, Suh, S.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
3U7L
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | (S)-N-(cyclopentylmethyl)-2-(3-(3,5-difluorophenyl)ureido)-N-(2-(hydroxyamino)-2-oxoethyl)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7M
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-4-(3-(2-fluorophenyl)ureido)-5-methyl-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3U7K
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | (S)-N-(cyclopentylmethyl)-N-(2-(hydroxyamino)-2-oxoethyl)-2-(3-(2-methoxyphenyl)ureido)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7N
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-5-methyl-4-(3-(5-methylpyridin-2-yl)ureido)-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3AKJ
 
 | |
3AKL
 
 | |
3AKK
 
 | |
4Y4V
 
 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUU
 
 | |
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1PX8
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
3NIQ
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase | | Descriptor: | 3-guanidinopropionase, GLYCEROL, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
